Overview of the Genetic Causes of Hereditary Breast and Ovarian Cancer Syndrome in a Large French Patient Cohort
- PMID: 37444530
- PMCID: PMC10341368
- DOI: 10.3390/cancers15133420
Overview of the Genetic Causes of Hereditary Breast and Ovarian Cancer Syndrome in a Large French Patient Cohort
Abstract
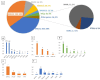
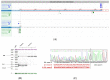
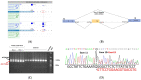
The use of multigene panel testing for patients with a predisposition to Hereditary Breast and Ovarian Cancer syndrome (HBOC) is increasing as the identification of mutations is useful for diagnosis and disease management. Here, we conducted a retrospective analysis of BRCA1/2 and non-BRCA gene sequencing in 4630 French HBOC suspected patients. Patients were investigated using a germline cancer panel including the 13 genes defined by The French Genetic and Cancer Group (GGC)-Unicancer. In the patients analyzed, 528 pathogenic and likely pathogenic variants (P/LP) were identified, including BRCA1 (n = 203, 38%), BRCA2 (n = 198, 37%), PALB2 (n = 46, 9%), RAD51C (n = 36, 7%), TP53 (n = 16, 3%), and RAD51D (n = 13, 2%). In addition, 35 novel (P/LP) variants, according to our knowledge, were identified, and double mutations in two distinct genes were found in five patients. Interestingly, retesting a subset of BRCA1/2-negative individuals with an expanded panel produced clinically relevant results in 5% of cases. Additionally, combining in silico (splicing impact prediction tools) and in vitro analyses (RT-PCR and Sanger sequencing) highlighted the deleterious impact of four candidate variants on splicing and translation. Our results present an overview of pathogenic variations of HBOC genes in the southeast of France, emphasizing the clinical relevance of cDNA analysis and the importance of retesting BRCA-negative individuals with an expanded panel.
Keywords: BRCA gene; HBOC; NGS; RNA analysis; multigene panel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Castéra L., Harter V., Muller E., Krieger S., Goardon N., Ricou A., Rousselin A., Paimparay G., Legros A., Bruet O., et al. Landscape of Pathogenic Variations in a Panel of 34 Genes and Cancer Risk Estimation from 5131 HBOC Families. Genet. Med. 2018;20:1677–1686. doi: 10.1038/s41436-018-0005-9. - DOI - PubMed
-
- Moretta J., Berthet P., Bonadona V., Caron O., Cohen-Haguenauer O., Colas C., Corsini C., Cusin V., De Pauw A., Delnatte C., et al. The French Genetic and Cancer Consortium guidelines for multigene panel analysis in hereditary breast and ovarian cancer predisposition. Bull. Cancer. 2018;105:907–917. doi: 10.1016/j.bulcan.2018.08.003. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

