Alterations in the Epigenetic Machinery Associated with Prostate Cancer Health Disparities
- PMID: 37444571
- PMCID: PMC10341375
- DOI: 10.3390/cancers15133462
Alterations in the Epigenetic Machinery Associated with Prostate Cancer Health Disparities
Abstract
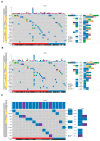
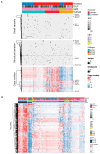
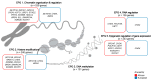
Prostate cancer is driven by acquired genetic alterations, including those impacting the epigenetic machinery. With African ancestry as a significant risk factor for aggressive disease, we hypothesize that dysregulation among the roughly 656 epigenetic genes may contribute to prostate cancer health disparities. Investigating prostate tumor genomic data from 109 men of southern African and 56 men of European Australian ancestry, we found that African-derived tumors present with a longer tail of epigenetic driver gene candidates (72 versus 10). Biased towards African-specific drivers (63 versus 9 shared), many are novel to prostate cancer (18/63), including several putative therapeutic targets (CHD7, DPF3, POLR1B, SETD1B, UBTF, and VPS72). Through clustering of all variant types and copy number alterations, we describe two epigenetic PCa taxonomies capable of differentiating patients by ancestry and predicted clinical outcomes. We identified the top genes in African- and European-derived tumors representing a multifunctional "generic machinery", the alteration of which may be instrumental in epigenetic dysregulation and prostate tumorigenesis. In conclusion, numerous somatic alterations in the epigenetic machinery drive prostate carcinogenesis, but African-derived tumors appear to achieve this state with greater diversity among such alterations. The greater novelty observed in African-derived tumors illustrates the significant clinical benefit to be derived from a much needed African-tailored approach to prostate cancer healthcare aimed at reducing prostate cancer health disparities.
Keywords: African ancestry; epigenetic machinery; epigenomics; health disparity; prostate cancer; somatic alteration; southern Africa.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lee W.H., Morton R.A., Epstein J.I., Brooks J.D., Campbell P.A., Bova G.S., Hsieh W.S., Isaacs W.B., Nelson W.G. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl. Acad. Sci. USA. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

