Improving Soluble Expression of SARS-CoV-2 Spike Priming Protease TMPRSS2 with an Artificial Fusing Protein
- PMID: 37445653
- PMCID: PMC10341623
- DOI: 10.3390/ijms241310475
Improving Soluble Expression of SARS-CoV-2 Spike Priming Protease TMPRSS2 with an Artificial Fusing Protein
Abstract
SARS-CoV-2 relies on the recognition of the spike protein by the host cell receptor ACE2 for cellular entry. In this process, transmembrane serine protease 2 (TMPRSS2) plays a pivotal role, as it acts as the principal priming agent catalyzing spike protein cleavage to initiate the fusion of the cell membrane with the virus. Thus, TMPRSS2 is an ideal pharmacological target for COVID-19 therapy development, and the effective production of high-quality TMPRSS2 protein is essential for basic and pharmacological research. Unfortunately, as a mammalian-originated protein, TMPRSS2 could not be solubly expressed in the prokaryotic system. In this study, we applied different protein engineering methods and found that an artificial protein XXA derived from an antifreeze protein can effectively promote the proper folding of TMPRSS2, leading to a significant improvement in the yield of its soluble form. Our study also showed that the fused XXA protein did not influence the enzymatic catalytic activity; instead, it greatly enhanced TMPRSS2's thermostability. Therefore, our strategy for increasing TMPRSS2 expression would be beneficial for the large-scale production of this stable enzyme, which would accelerate aniti-SARS-CoV-2 therapeutics development.
Keywords: SARS-CoV-2; TMPRSS2; artificial protein; protease; protein expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
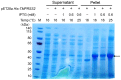
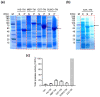
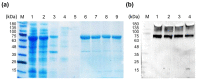

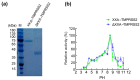
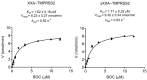
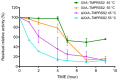
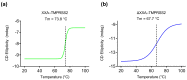
References
-
- Hoffmann M., Kleine–Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
-
- Papa G., Mallery D.L., Albecka A., Welch L.G., Cattin–Ortola J., Luptak J., Paul D., McMahon H.T., Goodfellow I.G., Carter A., et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell–cell fusion. PLoS Pathog. 2021;17:e1009246. doi: 10.1371/journal.ppat.1009246. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

