Physical Mapping of QTLs for Root Traits in a Population of Recombinant Inbred Lines of Hexaploid Wheat
- PMID: 37445670
- PMCID: PMC10341896
- DOI: 10.3390/ijms241310492
Physical Mapping of QTLs for Root Traits in a Population of Recombinant Inbred Lines of Hexaploid Wheat
Abstract
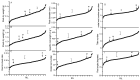
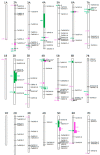
Root architecture is key in determining how effective plants are at intercepting and absorbing nutrients and water. Previously, the wheat (Triticum aestivum) cultivars Spica and Maringa were shown to have contrasting root morphologies. These cultivars were crossed to generate an F6:1 population of recombinant inbred lines (RILs) which was genotyped using a 90 K single nucleotide polymorphisms (SNP) chip. A total of 227 recombinant inbred lines (RILs) were grown in soil for 21 days in replicated trials under controlled conditions. At harvest, the plants were scored for seven root traits and two shoot traits. An average of 7.5 quantitative trait loci (QTL) were associated with each trait and, for each of these, physical locations of the flanking markers were identified using the Chinese Spring reference genome. We also compiled a list of genes from wheat and other monocotyledons that have previously been associated with root growth and morphology to determine their physical locations on the Chinese Spring reference genome. This allowed us to determine whether the QTL discovered in our study encompassed genes previously associated with root morphology in wheat or other monocotyledons. Furthermore, it allowed us to establish if the QTL were co-located with the QTL identified from previously published studies. The parental lines together with the genetic markers generated here will enable specific root traits to be introgressed into elite wheat lines. Moreover, the comprehensive list of genes associated with root development, and their physical locations, will be a useful resource for researchers investigating the genetics of root morphology in cereals.
Keywords: bread wheat; candidate genes; genetics; root morphology; traits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
QTLs Associated with Agronomic Traits in the Cutler × AC Barrie Spring Wheat Mapping Population Using Single Nucleotide Polymorphic Markers.PLoS One. 2016 Aug 11;11(8):e0160623. doi: 10.1371/journal.pone.0160623. eCollection 2016. PLoS One. 2016. PMID: 27513976 Free PMC article.
-
A SNP-based genetic dissection of versatile traits in bread wheat (Triticum aestivum L.).Plant J. 2021 Nov;108(4):960-976. doi: 10.1111/tpj.15407. Epub 2021 Nov 9. Plant J. 2021. PMID: 34218494
-
Prioritizing quantitative trait loci for root system architecture in tetraploid wheat.J Exp Bot. 2016 Feb;67(4):1161-78. doi: 10.1093/jxb/erw039. J Exp Bot. 2016. PMID: 26880749 Free PMC article.
-
Quick mapping and characterization of a co-located kernel length and thousand-kernel weight-related QTL in wheat.Theor Appl Genet. 2022 Aug;135(8):2849-2860. doi: 10.1007/s00122-022-04154-4. Epub 2022 Jul 8. Theor Appl Genet. 2022. PMID: 35804167 Review.
-
Regulation of root growth and elongation in wheat.Front Plant Sci. 2024 May 21;15:1397337. doi: 10.3389/fpls.2024.1397337. eCollection 2024. Front Plant Sci. 2024. PMID: 38835859 Free PMC article. Review.
Cited by
-
GA-sensitive Rht13 gene improves root architecture and osmotic stress tolerance in bread wheat.BMC Genom Data. 2024 Oct 24;25(1):90. doi: 10.1186/s12863-024-01272-4. BMC Genom Data. 2024. PMID: 39449141 Free PMC article.
References
-
- Kenobi K., Atkinson J.A., Wells D., Gaju O., De Silva J.G., Foulkes J., Dryden I.L., Wood A.T.A., Bennett M. Linear discriminant analysis reveals differences in root architecture in wheat seedlings related to nitrogen uptake efficiency. J. Exp. Bot. 2017;68:4969–4981. doi: 10.1093/jxb/erx300. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

