Virtual Screening Strategy and In Vitro Tests to Identify New Inhibitors of the Immunoproteasome
- PMID: 37445688
- PMCID: PMC10341482
- DOI: 10.3390/ijms241310504
Virtual Screening Strategy and In Vitro Tests to Identify New Inhibitors of the Immunoproteasome
Abstract
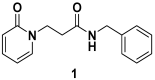
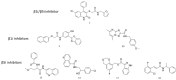
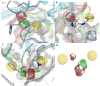
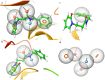
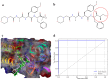
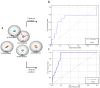
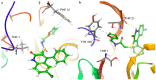
Immunoproteasome inhibition is a promising strategy for the treatment of hematological malignancies, autoimmune diseases, and inflammatory diseases. The design of non-covalent inhibitors of the immunoproteasome β1i/β5i catalytic subunits could be a novel approach to avoid the drawbacks of the known covalent inhibitors, such as toxicity due to off-target binding. In this work, we report the biological evaluation of thirty-four compounds selected from a commercially available collection. These hit compounds are the outcomes of a virtual screening strategy including a dynamic pharmacophore modeling approach onto the β1i subunit and a pharmacophore/docking approach onto the β5i subunit. The computational studies were first followed by in vitro enzymatic assays at 100 μM. Only compounds capable of inhibiting the enzymatic activity by more than 50% were characterized in detail using Tian continuous assays, determining the dissociation constant (Ki) of the non-covalent complex where Ki is also the measure of the binding affinity. Seven out of thirty-four hits showed to inhibit β1i and/or β5i subunit. Compound 3 is the most active on the β1i subunit with Ki = 11.84 ± 1.63 µM, and compound 17 showed Ki = 12.50 ± 0.77 µM on the β5i subunit. Compound 2 showed inhibitory activity on both subunits (Ki = 12.53 ± 0.18 and Ki = 31.95 ± 0.81 on the β1i subunit and β5i subunit, respectively). The induced fit docking analysis revealed interactions with Thr1 and Phe31 of β1i subunit and that represent new key residues as reported in our previous work. Onto β5i subunit, it interacts with the key residues Thr1, Thr21, and Tyr169. This last hit compound identified represents an interesting starting point for further optimization of β1i/β5i dual inhibitors of the immunoproteasome.
Keywords: docking; immunoproteasome; in vitro enzymatic assay; induced fit docking; pharmacophore modeling; β1i subunit; β5i subunit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Development of Novel Amides as Noncovalent Inhibitors of Immunoproteasomes.ChemMedChem. 2019 Apr 17;14(8):842-852. doi: 10.1002/cmdc.201900028. Epub 2019 Mar 26. ChemMedChem. 2019. PMID: 30829448
-
Identification of 2-thioxoimidazolidin-4-one derivatives as novel noncovalent proteasome and immunoproteasome inhibitors.Bioorg Med Chem Lett. 2018 Feb 1;28(3):278-283. doi: 10.1016/j.bmcl.2017.12.053. Epub 2017 Dec 26. Bioorg Med Chem Lett. 2018. PMID: 29292224
-
Fragment-Sized and Bidentate (Immuno)Proteasome Inhibitors Derived from Cysteine and Threonine Targeting Warheads.Cells. 2021 Dec 6;10(12):3431. doi: 10.3390/cells10123431. Cells. 2021. PMID: 34943940 Free PMC article.
-
The immunoproteasome in antigen processing and other immunological functions.Curr Opin Immunol. 2013 Feb;25(1):74-80. doi: 10.1016/j.coi.2012.11.004. Epub 2012 Dec 6. Curr Opin Immunol. 2013. PMID: 23219269 Review.
-
Immunoproteasome-Selective Inhibitors: A Promising Strategy to Treat Hematologic Malignancies, Autoimmune and Inflammatory Diseases.Curr Med Chem. 2016;23(12):1217-38. doi: 10.2174/0929867323666160318173706. Curr Med Chem. 2016. PMID: 26965184 Review.
Cited by
-
Computer-aided drug repurposing & discovery for Hepatitis B capsid protein.In Silico Pharmacol. 2025 Feb 25;13(1):35. doi: 10.1007/s40203-025-00314-8. eCollection 2025. In Silico Pharmacol. 2025. PMID: 40018383
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

