Cross-Linking Mass Spectrometry on P-Glycoprotein
- PMID: 37445813
- PMCID: PMC10341432
- DOI: 10.3390/ijms241310627
Cross-Linking Mass Spectrometry on P-Glycoprotein
Abstract
The ABC transporter P-glycoprotein (Pgp) has been found to be involved in multidrug resistance in tumor cells. Lipids and cholesterol have a pivotal role in Pgp's conformations; however, it is often difficult to investigate it with conventional structural biology techniques. Here, we applied robust approaches coupled with cross-linking mass spectrometry (XL-MS), where the natural lipid environment remains quasi-intact. Two experimental approaches were carried out using different cross-linkers (i) on living cells, followed by membrane preparation and immunoprecipitation enrichment of Pgp, and (ii) on-bead, subsequent to membrane preparation and immunoprecipitation. Pgp-containing complexes were enriched employing extracellular monoclonal anti-Pgp antibodies on magnetic beads, followed by on-bead enzymatic digestion. The LC-MS/MS results revealed mono-links on Pgp's solvent-accessible residues, while intraprotein cross-links confirmed a complex interplay between extracellular, transmembrane, and intracellular segments of the protein, of which several have been reported to be connected to cholesterol. Harnessing the MS results and those of molecular docking, we suggest an epitope for the 15D3 cholesterol-dependent mouse monoclonal antibody. Additionally, enriched neighbors of Pgp prove the strong connection of Pgp to the cytoskeleton and other cholesterol-regulated proteins. These findings suggest that XL-MS may be utilized for protein structure and network analyses in such convoluted systems as membrane proteins.
Keywords: P-glycoprotein; cancer; cholesterol; cross-linking mass spectrometry; membrane; mono-link; multidrug resistance; protein structure.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
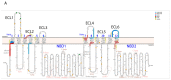




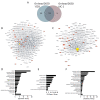


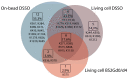
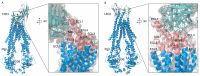
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

