The Influence of Lead and Acyrthosiphon pisum (Harris) on Generation of Pisum sativum Defense Signaling Molecules and Expression of Genes Involved in Their Biosynthesis
- PMID: 37445848
- PMCID: PMC10341517
- DOI: 10.3390/ijms241310671
The Influence of Lead and Acyrthosiphon pisum (Harris) on Generation of Pisum sativum Defense Signaling Molecules and Expression of Genes Involved in Their Biosynthesis
Abstract
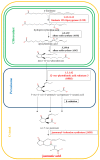
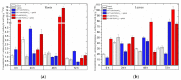
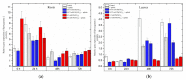
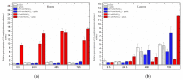
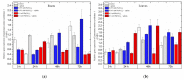
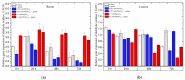
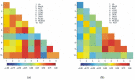
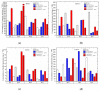
The main aim of this study was to understand the regulation of the biosynthesis of phytohormones as signaling molecules in the defense mechanisms of pea seedlings during the application of abiotic and biotic stress factors. It was important to identify this regulation at the molecular level in Pisum sativum L. seedlings under the influence of various concentrations of lead-i.e., a low concentration increasing plant metabolism, causing a hormetic effect, and a high dose causing a sublethal effect-and during feeding of a phytophagous insect with a piercing-sucking mouthpart-i.e., pea aphid (Acyrthosiphon pisum (Harris)). The aim of the study was to determine the expression level of genes encoding enzymes of the biosynthesis of signaling molecules such as phytohormones-i.e., jasmonates (JA/MeJA), ethylene (ET) and abscisic acid (ABA). Real-time qPCR was applied to analyze the expression of genes encoding enzymes involved in the regulation of the biosynthesis of JA/MeJA (lipoxygenase 1 (LOX1), lipoxygenase 2 (LOX2), 12-oxophytodienoate reductase 1 (OPR1) and jasmonic acid-amido synthetase (JAR1)), ET (1-aminocyclopropane-1-carboxylate synthase 3 (ACS3)) and ABA (9-cis-epoxycarotenoid dioxygenase (NCED) and aldehyde oxidase 1 (AO1)). In response to the abovementioned stress factors-i.e., abiotic and biotic stressors acting independently or simultaneously-the expression of the LOX1, LOX2, OPR1, JAR1, ACS3, NCED and AO1 genes at both sublethal and hormetic doses increased. Particularly high levels of the relative expression of the tested genes in pea seedlings growing at sublethal doses of lead and colonized by A. pisum compared to the control were noticeable. A hormetic dose of lead induced high expression levels of the JAR1, OPR1 and ACS3 genes, especially in leaves. Moreover, an increase in the concentration of phytohormones such as jasmonates (JA and MeJA) and aminococyclopropane-1-carboxylic acid (ACC)-ethylene (ET) precursor was observed. The results of this study indicate that the response of pea seedlings to lead and A. pisum aphid infestation differed greatly at both the gene expression and metabolic levels. The intensity of these defense responses depended on the organ, the metal dose and direct contact of the stress factor with the organ.
Keywords: Pisum sativum; ethylene; genes encoding enzymes of phytohormone biosynthesis; jasmonates; lead; pea aphid.
Conflict of interest statement
The authors declare no conflict of interests.
Figures




Similar articles
-
The Effects of Lead and Cross-Talk Between Lead and Pea Aphids on Defence Responses of Pea Seedlings.Int J Mol Sci. 2024 Nov 2;25(21):11804. doi: 10.3390/ijms252111804. Int J Mol Sci. 2024. PMID: 39519355 Free PMC article.
-
Differential induction of Pisum sativum defense signaling molecules in response to pea aphid infestation.Plant Sci. 2014 May;221-222:1-12. doi: 10.1016/j.plantsci.2014.01.011. Epub 2014 Jan 31. Plant Sci. 2014. PMID: 24656330
-
The Influence of Lead on Generation of Signalling Molecules and Accumulation of Flavonoids in Pea Seedlings in Response to Pea Aphid Infestation.Molecules. 2017 Aug 24;22(9):1404. doi: 10.3390/molecules22091404. Molecules. 2017. PMID: 28837107 Free PMC article.
-
Current understanding on ethylene signaling in plants: the influence of nutrient availability.Plant Physiol Biochem. 2013 Dec;73:128-38. doi: 10.1016/j.plaphy.2013.09.011. Epub 2013 Sep 20. Plant Physiol Biochem. 2013. PMID: 24095919 Review.
-
Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses.Front Plant Sci. 2015 Dec 1;6:1077. doi: 10.3389/fpls.2015.01077. eCollection 2015. Front Plant Sci. 2015. PMID: 26648959 Free PMC article. Review.
Cited by
-
Expression Profiling Analysis of the SWEET Gene Family in In Vitro Pitaya Under Low-Temperature Stress and Study of Its Cold Resistance Mechanism.Plants (Basel). 2024 Nov 2;13(21):3092. doi: 10.3390/plants13213092. Plants (Basel). 2024. PMID: 39520008 Free PMC article.
-
Effects of Jasmonic Acid on Stress Response and Quality Formation in Vegetable Crops and Their Underlying Molecular Mechanisms.Plants (Basel). 2024 Jun 4;13(11):1557. doi: 10.3390/plants13111557. Plants (Basel). 2024. PMID: 38891365 Free PMC article. Review.
-
The Effects of Lead and Cross-Talk Between Lead and Pea Aphids on Defence Responses of Pea Seedlings.Int J Mol Sci. 2024 Nov 2;25(21):11804. doi: 10.3390/ijms252111804. Int J Mol Sci. 2024. PMID: 39519355 Free PMC article.
-
The F-Box Protein TaFBA1 Positively Regulates Drought Resistance and Yield Traits in Wheat.Plants (Basel). 2024 Sep 16;13(18):2588. doi: 10.3390/plants13182588. Plants (Basel). 2024. PMID: 39339563 Free PMC article.
References
-
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. - DOI - PubMed
-
- Simmons B.I., Blyth P.S.A., Blanchard J.L., Clegg T., Delmas E., Garnier A., Griffiths C.A., Jacob U., Pennekamp F., Petchey O.L., et al. Refocusing multiple stressor research around the targets and scales of ecological impacts. Nat. Ecol. Evol. 2021;5:1478–1489. doi: 10.1038/s41559-021-01547-4. - DOI - PubMed
-
- Folt C.L., Chen C.Y., Moore M.V., Burnaford J. Synergism and antagonism among multiple stressors. Limnol. Oceanogr. 1999;44:864–877. doi: 10.4319/lo.1999.44.3_part_2.0864. - DOI
-
- Mai V.C., Drzewiecka K., Jeleń H., Narożna D., Rucińska-Sobkowiak R., Kęsy J., Floryszak-Wieczorek J., Gabryś B., Morkunas I. Differential induction of Pisum sativum defense signaling molecules in response to pea aphid infestation. Plant Sci. 2014;221:1–12. doi: 10.1016/j.plantsci.2014.01.011. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

