Transcriptome-Wide Identification of the GRAS Transcription Factor Family in Pinus massoniana and Its Role in Regulating Development and Stress Response
- PMID: 37445868
- PMCID: PMC10341488
- DOI: 10.3390/ijms241310690
Transcriptome-Wide Identification of the GRAS Transcription Factor Family in Pinus massoniana and Its Role in Regulating Development and Stress Response
Abstract
Pinus massoniana is a species used in afforestation and has high economic, ecological, and therapeutic significance. P. massoniana experiences a variety of biotic and abiotic stresses, and thus presents a suitable model for studying how woody plants respond to such stress. Numerous families of transcription factors are involved in the research of stress resistance, with the GRAS family playing a significant role in plant development and stress response. Though GRASs have been well explored in various plant species, much research remains to be undertaken on the GRAS family in P. massoniana. In this study, 21 PmGRASs were identified in the P. massoniana transcriptome. P. massoniana and Arabidopsis thaliana phylogenetic analyses revealed that the PmGRAS family can be separated into nine subfamilies. The results of qRT-PCR and transcriptome analyses under various stress and hormone treatments reveal that PmGRASs, particularly PmGRAS9, PmGRAS10 and PmGRAS17, may be crucial for stress resistance. The majority of PmGRASs were significantly expressed in needles and may function at multiple locales and developmental stages, according to tissue-specific expression analyses. Furthermore, the DELLA subfamily members PmGRAS9 and PmGRAS17 were nuclear localization proteins, while PmGRAS9 demonstrated transcriptional activation activity in yeast. The results of this study will help explore the relevant factors regulating the development of P. massoniana, improve stress resistance and lay the foundation for further identification of the biological functions of PmGRASs.
Keywords: GRAS; Pinus massoniana; abiotic stresses; development; expression; hormone treatments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




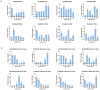


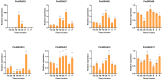

References
-
- Wang J.C., Qiu W.M., Jin K.M., Lu Z.C., Han X.J., Zhuo R.Y., Liu X.G., He Z.Q. Comprehensive analysis of WRKY gene family in Sedum plumbizincicola responding to cadmium stress. J. Nanjing For. Univ. Nat. 2023;47:49–60. doi: 10.12302/j.issn.1000-2006.202201015. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

