A New Method to Detect Variants of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification Combined with a Bioluminescent Assay in Real Time (RT-LAMP-BART)
- PMID: 37445876
- PMCID: PMC10341676
- DOI: 10.3390/ijms241310698
A New Method to Detect Variants of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification Combined with a Bioluminescent Assay in Real Time (RT-LAMP-BART)
Abstract
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), of which there are several variants. The three major variants (Alpha, Delta, and Omicron) carry the N501Y, L452R, and Q493R/Q498R mutations, respectively, in the S gene. Control of COVID-19 requires rapid and reliable detection of not only SARS-CoV-2 but also its variants. We previously developed a reverse transcription loop-mediated isothermal amplification assay combined with a bioluminescent assay in real time (RT-LAMP-BART) to detect the L452R mutation in the SARS-CoV-2 spike protein. In this study, we established LAMP primers and peptide nucleic acid probes to detect N501Y and Q493R/Q498R. The LAMP primer sets and PNA probes were designed for the N501Y and Q493R/Q498R mutations on the S gene of SARS-CoV-2. The specificities of RT-LAMP-BART assays were evaluated using five viral and four bacterial reference strains. The sensitivities of RT-LAMP-BART assays were evaluated using synthetic RNAs that included the target sequences, together with RNA-spiked clinical nasopharyngeal and salivary specimens. The results were compared with those of conventional real-time reverse transcription-polymerase chain reaction (RT-PCR) methods. The method correctly identified N501Y and Q493R/Q498R. Within 30 min, the RT-LAMP-BART assays detected up to 100-200 copies of the target genes; conventional real-time RT-PCR required 130 min and detected up to 500-3000 copies. Surprisingly, the real-time RT-PCR for N501Y did not detect the BA.1 and BA.2 variants (Omicron) that exhibited the N501Y mutation. The novel RT-LAMP-BART assay is highly specific and more sensitive than conventional real-time RT-PCR. The new assay is simple, inexpensive, and rapid; thus, it can be useful in efforts to identify SARS-CoV-2 variants of concern.
Keywords: SARS-CoV-2; loop-mediated isothermal amplification; spike protein; variants of concern.
Conflict of interest statement
L.T. and N.P. are employed by Erba Molecular and 3M Company, respectively. It did not influence the study design; collection, analysis, or interpretation of data; the writing of this article; or the decision to submit it for publication. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
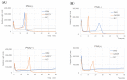
References
-
- Baek Y.H., Um J., Antigua K.J.C., Park J.H., Kim Y., Oh S., Kim Y.I., Choi W.S., Kim S.G., Jeong J.H., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. - DOI - PMC - PubMed
-
- Hodcroft E. Shared Mutations. [(accessed on 3 November 2022)]. Available online: https://covariants.org/shared-mutations.
-
- TAKARA about Detection of Omicron Strain (B.1.1.529+BA.* Strain) Using this Product Series. [(accessed on 1 April 2023)]. Available online: https://catalog.takara-bio.co.jp/product/basic_info.php?unitid=U100009522.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

