Extensive Bioinformatics Analyses Reveal a Phylogenetically Conserved Winged Helix (WH) Domain (Zτ) of Topoisomerase IIα, Elucidating Its Very High Affinity for Left-Handed Z-DNA and Suggesting Novel Putative Functions
- PMID: 37445918
- PMCID: PMC10341724
- DOI: 10.3390/ijms241310740
Extensive Bioinformatics Analyses Reveal a Phylogenetically Conserved Winged Helix (WH) Domain (Zτ) of Topoisomerase IIα, Elucidating Its Very High Affinity for Left-Handed Z-DNA and Suggesting Novel Putative Functions
Abstract
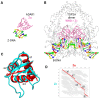
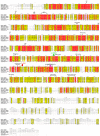
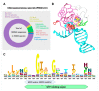
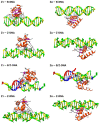
The dynamic processes operating on genomic DNA, such as gene expression and cellular division, lead inexorably to topological challenges in the form of entanglements, catenanes, knots, "bubbles", R-loops, and other outcomes of supercoiling and helical disruption. The resolution of toxic topological stress is the function attributed to DNA topoisomerases. A prominent example is the negative supercoiling (nsc) trailing processive enzymes such as DNA and RNA polymerases. The multiple equilibrium states that nscDNA can adopt by redistribution of helical twist and writhe include the left-handed double-helical conformation known as Z-DNA. Thirty years ago, one of our labs isolated a protein from Drosophila cells and embryos with a 100-fold greater affinity for Z-DNA than for B-DNA, and identified it as topoisomerase II (gene Top2, orthologous to the human UniProt proteins TOP2A and TOP2B). GTP increased the affinity and selectivity for Z-DNA even further and also led to inhibition of the isomerase enzymatic activity. An allosteric mechanism was proposed, in which topoII acts as a Z-DNA-binding protein (ZBP) to stabilize given states of topological (sub)domains and associated multiprotein complexes. We have now explored this possibility by comprehensive bioinformatic analyses of the available protein sequences of topoII representing organisms covering the whole tree of life. Multiple alignment of these sequences revealed an extremely high level of evolutionary conservation, including a winged-helix protein segment, here denoted as Zτ, constituting the putative structural homolog of Zα, the canonical Z-DNA/Z-RNA binding domain previously identified in the interferon-inducible RNA Adenosine-to-Inosine-editing deaminase, ADAR1p150. In contrast to Zα, which is separate from the protein segment responsible for catalysis, Zτ encompasses the active site tyrosine of topoII; a GTP-binding site and a GxxG sequence motif are in close proximity. Quantitative Zτ-Zα similarity comparisons and molecular docking with interaction scoring further supported the "B-Z-topoII hypothesis" and has led to an expanded mechanism for topoII function incorporating the recognition of Z-DNA segments ("Z-flipons") as an inherent and essential element. We further propose that the two Zτ domains of the topoII homodimer exhibit a single-turnover "conformase" activity on given G(ate) B-DNA segments ("Z-flipins"), inducing their transition to the left-handed Z-conformation. Inasmuch as the topoII-Z-DNA complexes are isomerase inactive, we infer that they fulfill important structural roles in key processes such as mitosis. Topoisomerases are preeminent targets of anti-cancer drug discovery, and we anticipate that detailed elucidation of their structural-functional interactions with Z-DNA and GTP will facilitate the design of novel, more potent and selective anti-cancer chemotherapeutic agents.
Keywords: GTP; Z-DNA; bioinformatics; topoII; topoisomerase IIα.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Zα and Zβ Localize ADAR1 to Flipons That Modulate Innate Immunity, Alternative Splicing, and Nonsynonymous RNA Editing.Int J Mol Sci. 2025 Mar 7;26(6):2422. doi: 10.3390/ijms26062422. Int J Mol Sci. 2025. PMID: 40141064 Free PMC article. Review.
-
Protein-induced B-Z transition of DNA duplex containing a 2'-OMe guanosine.Biochem Biophys Res Commun. 2020 Dec 10;533(3):417-423. doi: 10.1016/j.bbrc.2020.09.017. Epub 2020 Sep 21. Biochem Biophys Res Commun. 2020. PMID: 32972754
-
NMR investigation on the DNA binding and B-Z transition pathway of the Zα domain of human ADAR1.Biophys Chem. 2013 Feb;172:18-25. doi: 10.1016/j.bpc.2012.12.002. Epub 2012 Dec 21. Biophys Chem. 2013. PMID: 23334429
-
Thermodynamic analysis of Zα domain-nucleic acid interactions.Biochem J. 2022 Aug 31;479(16):1727-1741. doi: 10.1042/BCJ20220200. Biochem J. 2022. PMID: 35969150
-
Zalpha-domains: at the intersection between RNA editing and innate immunity.Semin Cell Dev Biol. 2012 May;23(3):275-80. doi: 10.1016/j.semcdb.2011.11.001. Epub 2011 Nov 7. Semin Cell Dev Biol. 2012. PMID: 22085847 Review.
Cited by
-
No Time to Relax and Unwind: Exploration of Topoisomerases and a Growing Field of Study.Int J Mol Sci. 2023 Aug 23;24(17):13080. doi: 10.3390/ijms241713080. Int J Mol Sci. 2023. PMID: 37685888 Free PMC article.
-
Zα and Zβ Localize ADAR1 to Flipons That Modulate Innate Immunity, Alternative Splicing, and Nonsynonymous RNA Editing.Int J Mol Sci. 2025 Mar 7;26(6):2422. doi: 10.3390/ijms26062422. Int J Mol Sci. 2025. PMID: 40141064 Free PMC article. Review.
-
Flipons and small RNAs accentuate the asymmetries of pervasive transcription by the reset and sequence-specific microcoding of promoter conformation.J Biol Chem. 2023 Sep;299(9):105140. doi: 10.1016/j.jbc.2023.105140. Epub 2023 Aug 5. J Biol Chem. 2023. PMID: 37544644 Free PMC article. Review.
References
-
- Wang J.C. Untangling the Double Helix: DNA Entanglement and the Action of the DNA Topoisomerases. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2009.
-
- Neidle S., Sanderson M. Principles of Nucleic Acid Structure. 2nd ed. Academic Press; Cambridge, MA, USA: 2021.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

