Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids
- PMID: 37445941
- PMCID: PMC10341749
- DOI: 10.3390/ijms241310763
Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids
Abstract
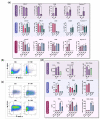
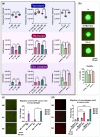
Reprogramming of tumor-associated macrophages (TAMs) is a promising strategy for cancer immunotherapy. Several studies have shown that cancer cells induce/support the formation of immunosuppressive TAMs phenotypes. However, the specific factors that orchestrate this immunosuppressive process are unknown or poorly studied. In vivo studies are expensive, complex, and ethically constrained. Therefore, 3D cell interaction models could become a unique framework for the identification of important TAMs programming factors. In this study, we have established and characterized a new in vitro 3D model for macrophage programming in the presence of cancer cell spheroids. First, it was demonstrated that the profile of cytokines, chemokines, and surface markers of 3D-cultured macrophages did not differ conceptually from monolayer-cultured M1 and M2-programmed macrophages. Second, the possibility of reprogramming macrophages in 3D conditions was investigated. In total, the dynamic changes in 6 surface markers, 11 cytokines, and 22 chemokines were analyzed upon macrophage programming (M1 and M2) and reprogramming (M1→M2 and M2→M1). According to the findings, the reprogramming resulted in a mixed macrophage phenotype that expressed both immunosuppressive and anti-cancer immunostimulatory features. Third, cancer cell spheroids were shown to stimulate the production of immunosuppressive M2 markers as well as pro-tumor cytokines and chemokines. In summary, the newly developed 3D model of cancer cell spheroid/macrophage co-culture under free-floating conditions can be used for studies on macrophage plasticity and for the development of targeted cancer immunotherapy.
Keywords: 3D cell culture models; cancer cell spheroids; chemokines; cytokines; macrophage plasticity; macrophage polarization; macrophage reprogramming; tumor-associated macrophages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

