Yeast Heterologous Expression Systems for the Study of Plant Membrane Proteins
- PMID: 37445944
- PMCID: PMC10342074
- DOI: 10.3390/ijms241310768
Yeast Heterologous Expression Systems for the Study of Plant Membrane Proteins
Abstract
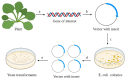
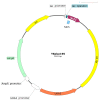
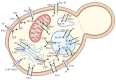
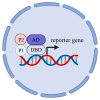
Researchers are often interested in proteins that are present in cells in small ratios compared to the total amount of proteins. These proteins include transcription factors, hormones and specific membrane proteins. However, sufficient amounts of well-purified protein preparations are required for functional and structural studies of these proteins, including the creation of artificial proteoliposomes and the growth of protein 2D and 3D crystals. This aim can be achieved by the expression of the target protein in a heterologous system. This review describes the applications of yeast heterologous expression systems in studies of plant membrane proteins. An initial brief description introduces the widely used heterologous expression systems of the baker's yeast Saccharomyces cerevisiae and the methylotrophic yeast Pichia pastoris. S. cerevisiae is further considered a convenient model system for functional studies of heterologously expressed proteins, while P. pastoris has the advantage of using these yeast cells as factories for producing large quantities of proteins of interest. The application of both expression systems is described for functional and structural studies of membrane proteins from plants, namely, K+- and Na+-transporters, various ATPases and anion transporters, and other transport proteins.
Keywords: Pichia pastoris; Saccharomyces cerevisiae; baker’s yeasts; heterologous expression; methylotrophic yeasts; plant membrane proteins; recombinant proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

