Comparative Transcriptome Analysis Reveals Novel Candidate Resistance Genes Involved in Defence against Phytophthora cactorum in Strawberry
- PMID: 37446029
- PMCID: PMC10341869
- DOI: 10.3390/ijms241310851
Comparative Transcriptome Analysis Reveals Novel Candidate Resistance Genes Involved in Defence against Phytophthora cactorum in Strawberry
Abstract
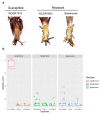
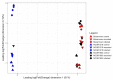
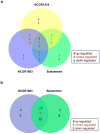
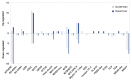
Crown rot, caused by Phytophthora cactorum, is a devastating disease of strawberry. While most commercial octoploid strawberry cultivars (Fragaria × ananassa Duch) are generally susceptible, the diploid species Fragaria vesca is a potential source of resistance genes to P. cactorum. We previously reported several F. vesca genotypes with varying degrees of resistance to P. cactorum. To gain insights into the strawberry defence mechanisms, comparative transcriptome profiles of two resistant genotypes (NCGR1603 and Bukammen) and a susceptible genotype (NCGR1218) of F. vesca were analysed by RNA-Seq after wounding and subsequent inoculation with P. cactorum. Differential gene expression analysis identified several defence-related genes that are highly expressed in the resistant genotypes relative to the susceptible genotype in response to P. cactorum after wounding. These included putative disease resistance (R) genes encoding receptor-like proteins, receptor-like kinases, nucleotide-binding sites, leucine-rich repeat proteins, RPW8-type disease resistance proteins, and 'pathogenesis-related protein 1'. Seven of these R-genes were expressed only in the resistant genotypes and not in the susceptible genotype, and these appeared to be present only in the genomes of the resistant genotypes, as confirmed by PCR analysis. We previously reported a single major gene locus RPc-1 (Resistance to Phytophthora cactorum 1) in F. vesca that contributed resistance to P. cactorum. Here, we report that 4-5% of the genes (35-38 of ca 800 genes) in the RPc-1 locus are differentially expressed in the resistant genotypes compared to the susceptible genotype after inoculation with P. cactorum. In particular, we identified three defence-related genes encoding wall-associated receptor-like kinase 3, receptor-like protein 12, and non-specific lipid-transfer protein 1-like that were highly expressed in the resistant genotypes compared to the susceptible one. The present study reports several novel candidate disease resistance genes that warrant further investigation for their role in plant defence against P. cactorum.
Keywords: Fragaria vesca; crown rot; disease resistance (R-genes); oomycete.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Reprogramming of Strawberry (Fragaria vesca) Root Transcriptome in Response to Phytophthora cactorum.PLoS One. 2016 Aug 12;11(8):e0161078. doi: 10.1371/journal.pone.0161078. eCollection 2016. PLoS One. 2016. PMID: 27518577 Free PMC article.
-
Expression of resistance gene analogs in woodland strawberry (Fragaria vesca) during infection with Phytophthora cactorum.Mol Genet Genomics. 2016 Oct;291(5):1967-78. doi: 10.1007/s00438-016-1232-x. Epub 2016 Jul 22. Mol Genet Genomics. 2016. PMID: 27447867 Free PMC article.
-
Terpenoid and lipid profiles vary in different Phytophthora cactorum - strawberry interactions.Phytochemistry. 2021 Sep;189:112820. doi: 10.1016/j.phytochem.2021.112820. Epub 2021 Jun 3. Phytochemistry. 2021. PMID: 34091112
-
Quantitative trait loci controlling Phytophthora cactorum resistance in the cultivated octoploid strawberry (Fragaria × ananassa).Hortic Res. 2019 May 1;6:60. doi: 10.1038/s41438-019-0136-4. eCollection 2019. Hortic Res. 2019. PMID: 31069084 Free PMC article.
-
Characterization of expansin genes and their transcriptional regulation by histone modifications in strawberry.Planta. 2021 Jul 3;254(2):21. doi: 10.1007/s00425-021-03665-6. Planta. 2021. PMID: 34216276 Review.
Cited by
-
Transcriptome analysis of Phytophthora cactorum infecting strawberry identified RXLR effectors that induce cell death when transiently expressed in Nicotiana benthamiana.Front Plant Sci. 2024 May 24;15:1379970. doi: 10.3389/fpls.2024.1379970. eCollection 2024. Front Plant Sci. 2024. PMID: 38855473 Free PMC article.
-
Transcriptome Analysis Reveals the Molecular Mechanisms of Carrot Adaptation to Alternaria Leaf Blight.Int J Mol Sci. 2024 Dec 6;25(23):13106. doi: 10.3390/ijms252313106. Int J Mol Sci. 2024. PMID: 39684815 Free PMC article.
-
Secondary Metabolites and Their Role in Strawberry Defense.Plants (Basel). 2023 Sep 12;12(18):3240. doi: 10.3390/plants12183240. Plants (Basel). 2023. PMID: 37765404 Free PMC article. Review.
-
Bioinformatics Analysis of the Panax ginseng Cyclophilin Gene and Its Anti-Phytophthora cactorum Activity.Plants (Basel). 2024 Sep 29;13(19):2731. doi: 10.3390/plants13192731. Plants (Basel). 2024. PMID: 39409601 Free PMC article.
References
-
- Golzar H., Phillips D., Mack S. Occurrence of Strawberry Root and Crown Rot in Western Australia. Australas. Plant Dis. Notes. 2007;2:145. doi: 10.1071/DN07057. - DOI
-
- Stensvand A., Herrero M.L., Talgø V. Crown Rot Caused by Phytophthora cactorum in Norwegian Strawberry Production. EPPO Bull. 1999;29:155–158. doi: 10.1111/j.1365-2338.1999.tb00809.x. - DOI
-
- van der Scheer H.A.T. Isolation of Phytophthora cactorum from Soil in Orchards and Strawberry Fields and Differences in Pathogenicity to Apple. Neth. J. Plant Pathol. 1971;77:65–72. doi: 10.1007/BF01981494. - DOI
-
- Hantula J., Lilja A., Nuorteva H., Parikka P., Werres S. Pathogenicity, Morphology and Genetic Variation of Phytophthora cactorum from Strawberry, Apple, Rhododendron, and Silver Birch. Mycol. Res. 2000;104:1062–1068. doi: 10.1017/S0953756200002999. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

