Genome-Wide Identification and Expression Analysis of ACTIN Family Genes in the Sweet Potato and Its Two Diploid Relatives
- PMID: 37446107
- PMCID: PMC10341734
- DOI: 10.3390/ijms241310930
Genome-Wide Identification and Expression Analysis of ACTIN Family Genes in the Sweet Potato and Its Two Diploid Relatives
Abstract
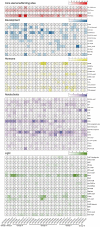
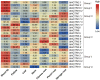
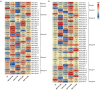
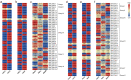
ACTINs are structural proteins widely distributed in plants. They are the main components of microfilaments and participate in many crucial physiological activities, including the maintenance of cell shape and cytoplasmic streaming. Meanwhile, ACTIN, as a housekeeping gene, is widely used in qRT-PCR analyses of plants. However, ACTIN family genes have not been explored in the sweet potato. In this study, we identified 30, 39, and 44 ACTINs in the cultivated hexaploid sweet potato (Ipomoea batatas, 2n = 6x = 90) and its two diploid relatives, Ipomoea trifida (2n = 2x = 30) and Ipomoea triloba (2n = 2x = 30), respectively, via analysis of their genome structure and by phylogenetic characterization. These ACTINs were divided into six subgroups according to their phylogenetic relationships with Arabidopsis thaliana. The physiological properties of the protein, chromosome localization, phylogenetic relationship, gene structure, promoter cis-elements, protein interaction networks, and expression patterns of these 113 ACTINs were systematically investigated. The results suggested that homologous ACTINs are differentiated in the sweet potato and its two diploid relatives, and play various vital roles in plant growth, tuberous root development, hormone crosstalk, and abiotic stress responses. Some stable ACTINs that could be used as internal reference genes were found in the sweet potato and its two diploid relatives, e.g., IbACTIN18, -20, and -16.2; ItfACTIN2.2, -16, and -10; ItbACTIN18 and -19.1. This work provides a comprehensive comparison and furthers our understanding of the ACTIN genes in the sweet potato and its two diploid relatives, thereby supplying a theoretical foundation for their functional study and further facilitating the molecular breeding of sweet potatoes.
Keywords: ACTIN; I. trifida; I. triloba; abiotic stress; hormone treatment; sweet potato; tissue-specific expression; tuberous root development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Baluska F., Hlavacka A., Samaj J., Palme K., Robinson D.G., Matoh T., McCurdy D.W., Menzel D., Volkmann D. F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol. 2002;130:422–431. doi: 10.1104/pp.007526. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

