Genome-Wide Identification of MADS-Box Genes in Taraxacum kok-saghyz and Taraxacum mongolicum: Evolutionary Mechanisms, Conserved Functions and New Functions Related to Natural Rubber Yield Formation
- PMID: 37446175
- PMCID: PMC10341699
- DOI: 10.3390/ijms241310997
Genome-Wide Identification of MADS-Box Genes in Taraxacum kok-saghyz and Taraxacum mongolicum: Evolutionary Mechanisms, Conserved Functions and New Functions Related to Natural Rubber Yield Formation
Abstract
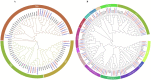
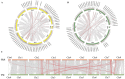
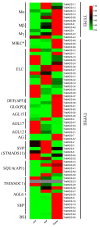
MADS-box transcription regulators play important roles in plant growth and development. However, very few MADS-box genes have been isolated in the genus Taraxacum, which consists of more than 3000 species. To explore their functions in the promising natural rubber (NR)-producing plant Taraxacum kok-saghyz (TKS), MADS-box genes were identified in the genome of TKS and the related species Taraxacum mongolicum (TM; non-NR-producing) via genome-wide screening. In total, 66 TkMADSs and 59 TmMADSs were identified in the TKS and TM genomes, respectively. From diploid TKS to triploid TM, the total number of MADS-box genes did not increase, but expansion occurred in specific subfamilies. Between the two genomes, a total of 11 duplications, which promoted the expansion of MADS-box genes, were identified in the two species. TkMADS and TmMADS were highly conserved, and showed good collinearity. Furthermore, most TkMADS genes exhibiting tissue-specific expression patterns, especially genes associated with the ABCDE model, were preferentially expressed in the flowers, suggesting their conserved and dominant functions in flower development in TKS. Moreover, by comparing the transcriptomes of different TKS lines, we identified 25 TkMADSs related to biomass formation and 4 TkMADSs related to NR content, which represented new targets for improving the NR yield of TKS.
Keywords: MADS-box gene; Taraxacum kok-saghyz; Taraxacum mongolicum; biomass; collinearity; natural rubber biosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., de Pouplana L.R., Martínez-Castilla L., Yanofsky M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

