Improving Rice Leaf Shape Using CRISPR/Cas9-Mediated Genome Editing of SRL1 and Characterizing Its Regulatory Network Involved in Leaf Rolling through Transcriptome Analysis
- PMID: 37446265
- PMCID: PMC10342453
- DOI: 10.3390/ijms241311087
Improving Rice Leaf Shape Using CRISPR/Cas9-Mediated Genome Editing of SRL1 and Characterizing Its Regulatory Network Involved in Leaf Rolling through Transcriptome Analysis
Abstract
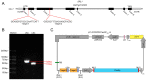
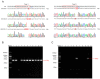
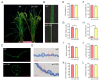
Leaf rolling is a crucial agronomic trait to consider in rice (Oryza sativa L.) breeding as it keeps the leaves upright, reducing interleaf shading and improving photosynthetic efficiency. The SEMI-ROLLED LEAF 1 (SRL1) gene plays a key role in regulating leaf rolling, as it encodes a glycosylphosphatidylinositol-anchored protein located on the plasma membrane. In this study, we used CRISPR/Cas9 to target the second and third exons of the SRL1 gene in the indica rice line GXU103, which resulted in the generation of 14 T0 transgenic plants with a double-target mutation rate of 21.4%. After screening 120 T1 generation plants, we identified 26 T-DNA-free homozygous double-target mutation plants. We designated the resulting SRL1 homozygous double-target knockout as srl1-103. This line exhibited defects in leaf development, leaf rolling in the mature upright leaves, and a compact nature of the fully grown plants. Compared with the wild type (WT), the T2 generation of srl1-103 varied in two key aspects: the width of flag leaf (12.6% reduction compared with WT) and the leaf rolling index (48.77% increase compared with WT). In order to gain a deeper understanding of the involvement of SRL1 in the regulatory network associated with rice leaf development, we performed a transcriptome analysis for the T2 generation of srl1-103. A comparison of srl1-103 with WT revealed 459 differentially expressed genes (DEGs), including 388 upregulated genes and 71 downregulated genes. In terms of the function of the DEGs, there seemed to be a significant enrichment of genes associated with cell wall synthesis (LOC_Os08g01670, LOC_Os05g46510, LOC_Os04g51450, LOC_Os10g28080, LOC_Os04g39814, LOC_Os01g71474, LOC_Os01g71350, and LOC_Os11g47600) and vacuole-related genes (LOC_Os09g23300), which may partially explain the increased leaf rolling in srl1-103. Furthermore, the significant downregulation of BAHD acyltransferase-like protein gene (LOC_Os08g44840) could be the main reason for the decreased leaf angle and the compact nature of the mutant plants. In summary, this study successfully elucidated the gene regulatory network in which SRL1 participates, providing theoretical support for targeting this gene in rice breeding programs to promote variety improvement.
Keywords: CRISPR/Cas9; SRL1; leaf rolling; rice; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells.Plant Physiol. 2012 Aug;159(4):1488-500. doi: 10.1104/pp.112.199968. Epub 2012 Jun 19. Plant Physiol. 2012. PMID: 22715111 Free PMC article.
-
CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice.Plant J. 2017 Dec;92(5):904-923. doi: 10.1111/tpj.13728. Epub 2017 Nov 8. Plant J. 2017. PMID: 28960566
-
CRISPR/Cas9 Guided Mutagenesis of Grain Size 3 Confers Increased Rice (Oryza sativa L.) Grain Length by Regulating Cysteine Proteinase Inhibitor and Ubiquitin-Related Proteins.Int J Mol Sci. 2021 Mar 22;22(6):3225. doi: 10.3390/ijms22063225. Int J Mol Sci. 2021. PMID: 33810044 Free PMC article.
-
CRISPR-Based Genome Editing: Advancements and Opportunities for Rice Improvement.Int J Mol Sci. 2022 Apr 18;23(8):4454. doi: 10.3390/ijms23084454. Int J Mol Sci. 2022. PMID: 35457271 Free PMC article. Review.
-
Improving yield-related traits by editing the promoter and distal regulatory region of heading date genes Ghd7 and PRR37 in elite rice variety Mei Xiang Zhan 2.Theor Appl Genet. 2025 Apr 5;138(4):92. doi: 10.1007/s00122-025-04880-5. Theor Appl Genet. 2025. PMID: 40186758 Review.
Cited by
-
Advances in Molecular Plant Sciences.Int J Mol Sci. 2024 Jun 10;25(12):6408. doi: 10.3390/ijms25126408. Int J Mol Sci. 2024. PMID: 38928115 Free PMC article.
References
-
- Ma H., Chong K., Deng X.W. Rice research: Past, present and future. J. Integr. Plant Biol. 2007;49:729. doi: 10.1111/j.1744-7909.2007.00515.x. - DOI
-
- Li L., Xue X., Chen Z., Zhang Y., Ma Y., Pan C., Zhu J., Pan X., Zuo S. Isolation and characterization of rl (t), a gene that controls leaf rolling in rice. Chin. Sci. Bull. 2014;59:3142–3152. doi: 10.1007/s11434-014-0357-8. - DOI
-
- Chen Z., Zuo S., Zhang Y., Li L., Pan X., Ma Y. Current progress in genetics research and breeding application of rolled leaf in rice. J. Yangzhou Univ. 2010;31:22–27. doi: 10.16872/j.cnki.1671-4652.2010.04.005. - DOI
-
- Cheng S.H., Cao L.Y., Zhuang J.Y., Chen S.G., Zhan X.D., Fan Y.Y., Zhu D.F., Min S.K. Super Hybrid Rice Breeding in China: Achievements and Prospects. J. Integr. Plant Biol. 2007;49:805–810. doi: 10.1111/j.1744-7909.2007.00514.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

