High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1
- PMID: 37446375
- PMCID: PMC10342360
- DOI: 10.3390/ijms241311197
High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1
Abstract
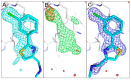
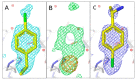
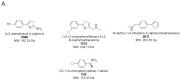
The identification of multiple simultaneous orientations of small molecule inhibitors binding to a protein target is a common challenge. It has recently been reported that the conformational heterogeneity of ligands is widely underreported in the Protein Data Bank, which is likely to impede optimal exploitation to improve affinity of these ligands. Significantly less is even known about multiple binding orientations for fragments (<300 Da), although this information would be essential for subsequent fragment optimisation using growing, linking or merging and rational structure-based design. Here, we use recently reported fragment hits for the SARS-CoV-2 non-structural protein 1 (nsp1) N-terminal domain to propose a general procedure for unambiguously identifying binding orientations of 2-dimensional fragments containing either sulphur or chloro substituents within the wavelength range of most tunable beamlines. By measuring datasets at two energies, using a tunable beamline operating in vacuum and optimised for data collection at very low X-ray energies, we show that the anomalous signal can be used to identify multiple orientations in small fragments containing sulphur and/or chloro substituents or to verify recently reported conformations. Although in this specific case we identified the positions of sulphur and chlorine in fragments bound to their protein target, we are confident that this work can be further expanded to additional atoms or ions which often occur in fragments. Finally, our improvements in the understanding of binding orientations will also serve to improve the rational optimisation of SARS-CoV-2 nsp1 fragment hits.
Keywords: COVID-19; SARS-CoV-2; anomalous difference Fourier map; fragment orientation; non-structural proteins; nsp1; tunable wavelength.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.Acta Crystallogr D Struct Biol. 2024 Jun 1;80(Pt 6):451-463. doi: 10.1107/S2059798324004480. Epub 2024 Jun 5. Acta Crystallogr D Struct Biol. 2024. PMID: 38841886 Free PMC article.
-
Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.Int J Mol Sci. 2022 Oct 18;23(20):12448. doi: 10.3390/ijms232012448. Int J Mol Sci. 2022. PMID: 36293303 Free PMC article.
-
Modelling studies reveal the importance of the C-terminal inter motif loop of NSP1 as a promising target site for drug discovery and screening of potential phytochemicals to combat SARS-CoV-2.J Mol Graph Model. 2021 Jul;106:107920. doi: 10.1016/j.jmgm.2021.107920. Epub 2021 Apr 19. J Mol Graph Model. 2021. PMID: 33933885 Free PMC article.
-
Overview and new developments in softer X-ray (2A < lambda < 5A) protein crystallography.J Synchrotron Radiat. 2004 Jan 1;11(Pt 1):1-3. doi: 10.1107/s0909049503024099. Epub 2003 Nov 28. J Synchrotron Radiat. 2004. PMID: 14646119 Review.
-
SARS-CoV-2 Proteins: Are They Useful as Targets for COVID-19 Drugs and Vaccines?Curr Mol Med. 2022;22(1):50-66. doi: 10.2174/1566524021666210223143243. Curr Mol Med. 2022. PMID: 33622224 Review.
Cited by
-
High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.Acta Crystallogr D Struct Biol. 2024 Jun 1;80(Pt 6):451-463. doi: 10.1107/S2059798324004480. Epub 2024 Jun 5. Acta Crystallogr D Struct Biol. 2024. PMID: 38841886 Free PMC article.
-
SARS-CoV2 Nsp1 is a metal-dependent DNA and RNA endonuclease.Biometals. 2024 Oct;37(5):1127-1146. doi: 10.1007/s10534-024-00596-z. Epub 2024 Mar 28. Biometals. 2024. PMID: 38538957 Free PMC article.
References
-
- van Zundert G.C.P., Hudson B.M., de Oliveira S.H.P., Keedy D.A., Fonseca R., Heliou A., Suresh P., Borrelli K., Day T., Fraser J.S., et al. qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps. J. Med. Chem. 2018;61:11183–11198. doi: 10.1021/acs.jmedchem.8b01292. - DOI - PMC - PubMed
-
- Deschamps J.R. The Role of Crystallography in Drug Design. In: Rapaka R.S., Sadée W., editors. Drug Addiction: From Basic Research to Therapy. Springer; New York, NY, USA: 2008. pp. 343–355. - DOI
-
- Han G.W., Lee J.Y., Song H.K., Chang C., Min K., Moon J., Shin D.H., Kopka M.L., Sawaya M.R., Yuan H.S., et al. Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography1 1Edited by D. Rees. J. Mol. Biol. 2001;308:263–278. doi: 10.1006/jmbi.2001.4559. - DOI - PubMed
-
- Murthy K.H., Winborne E.L., Minnich M.D., Culp J.S., Debouck C. The crystal structures at 2.2-A resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations. J. Biol. Chem. 1992;267:22770–22778. doi: 10.1016/S0021-9258(18)50014-4. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

