Broad-Spectrum, Potent, and Durable Ceria Nanoparticles Inactivate RNA Virus Infectivity by Targeting Virion Surfaces and Disrupting Virus-Receptor Interactions
- PMID: 37446852
- PMCID: PMC10343717
- DOI: 10.3390/molecules28135190
Broad-Spectrum, Potent, and Durable Ceria Nanoparticles Inactivate RNA Virus Infectivity by Targeting Virion Surfaces and Disrupting Virus-Receptor Interactions
Abstract
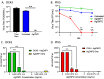
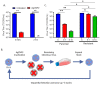
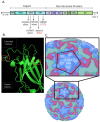
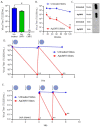
There is intense interest in developing long-lasting, potent, and broad-spectrum antiviral disinfectants. Ceria nanoparticles (CNPs) can undergo surface redox reactions (Ce3+ ↔ Ce4+) to generate ROS without requiring an external driving force. Here, we tested the mechanism behind our prior finding of potent inactivation of enveloped and non-enveloped RNA viruses by silver-modified CNPs, AgCNP1 and AgCNP2. Treatment of human respiratory viruses, coronavirus OC43 and parainfluenza virus type 5 (PIV5) with AgCNP1 and 2, respectively, prevented virus interactions with host cell receptors and resulted in virion aggregation. Rhinovirus 14 (RV14) mutants were selected to be resistant to inactivation by AgCNP2. Sequence analysis of the resistant virus genomes predicted two amino acid changes in surface-located residues D91V and F177L within capsid protein VP1. Consistent with the regenerative properties of CNPs, surface-applied AgCNP1 and 2 inactivated a wide range of structurally diverse viruses, including enveloped (OC43, SARS-CoV-2, and PIV5) and non-enveloped RNA viruses (RV14 and feline calicivirus; FCV). Remarkably, a single application of AgCNP1 and 2 potently inactivated up to four sequential rounds of virus challenge. Our results show broad-spectrum and long-lasting anti-viral activity of AgCNP nanoparticles, due to targeting of viral surface proteins to disrupt interactions with cellular receptors.
Keywords: anti-viral; coronavirus; disinfectant; nanoparticles; norovirus; parainfluenza; rhinovirus; virucidal.
Conflict of interest statement
C.D. commercialization of AgCNPs through Kismet Technologies.
Figures



References
-
- Knipe D.M., Howley P. Fields Virology. Volumes 1 and 2 LWW; Philadelphia, PA, USA: 2013.
-
- Ikonen N., Savolainen-Kopra C., Enstone J.E., Kulmala I., Pasanen P., Salmela A., Salo S., Nguyen-Van-Tam J.S., Ruutu P., PANDHUB consortium Deposition of respiratory virus pathogens on frequently touched surfaces at airports. BMC Infect. Dis. 2018;18:437. doi: 10.1186/s12879-018-3150-5. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

