Identification of Floral Volatile Components and Expression Analysis of Controlling Gene in Paeonia ostii 'Fengdan' under Different Cultivation Conditions
- PMID: 37447013
- PMCID: PMC10346646
- DOI: 10.3390/plants12132453
Identification of Floral Volatile Components and Expression Analysis of Controlling Gene in Paeonia ostii 'Fengdan' under Different Cultivation Conditions
Abstract
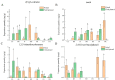
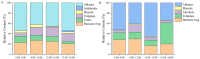
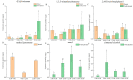
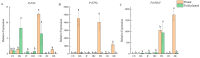
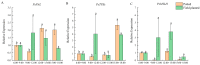
In order to explore the release rule of floral volatile substances and the diurnal variation of different flower development stages of Paeonia ostii 'Fengdan' in potted and ground-planted conditions, dynamic headspace adsorption combined with gas chromatography-mass spectrometry(GC-MS) was used to analyze the dynamic changes in floral volatile components and contents. Quantitative real-time PCR (qRT-PCR) was used to analyze changes in flower fragrance-regulating genes PsPAL, PsTPSs, and PsbHLH at different flower development stages and a daily change process at the full-blooming stage. The results show that there were differences in aroma components and contents of Paeonia ostii 'Fengdan' at different flower development stages and different time quantum of every day. There were 25 and 28 aroma components identified in 7 flower development stages of tree peonies planted in pots and in the field, respectively, and 23 and 22 aroma components identified at different time quantum of the day, of which the largest and highest content was alkanes. The main characteristic aroma substances were (E)-β-ocimene, 1,3,5-trimethoxybenzene, 2,4-di-tert-butylphenol, methyl jasmonate, nerol, and cinnamyl alcohol; released amounts of the abovementioned substances varied depending on the development stage and the time of the day. The expression of flower fragrance-controlling genes (PsPAL, PsTPSs, and PsbHLH) in tree peonies varied greatly in different conditions. The results of this study provide a valuable resource to investigate floral fragrance formation in tree peonies.
Keywords: Paeonia ostii; controlling gene; different cultivation conditions; floral volatile components.
Conflict of interest statement
We declare that we have no known competing financial interest or personal relationship that could have appeared to influence the work reported in this paper.
Figures

References
-
- Zhang L., Guo D.L., Guo L.L., Guo Q., Wang H.F., Hou X.G. Construction of a high-density genetic map and QTLs 410 mapping with GBS from the interspecific F1 population of P. ostii ‘Fengdan Bai’ and P. suffruticosa ‘Xin Riyuejin’. Sci. Hortic. 2019;246:190–200. doi: 10.1016/j.scienta.2018.10.039. - DOI
-
- Luo X.N., Sun D.Y., Wang S., Luo S., Fu Y.Q., Niu L.X., Shi Q.Q., Zhang Y.L. Integrating full-length transcriptomics and metabolomics reveals the regulatory mechanisms underlying yellow pigmentation in tree peony (Paeonia suffruticosa Andr.) flowers. Hortic. Res. 2021;8:235. doi: 10.1038/s41438-021-00666-0. - DOI - PMC - PubMed
-
- Guo L.L., Li Y.Y., Lei Y., Gao J.S., Song C.W., Guo D.L., Hou X.G. Genome-scale investigation and identification of variations associated with early flowering based on whole genome resequencing and transcriptome integrated analysis in tree peony. Sci. Hortic. 2023;310:111695. doi: 10.1016/j.scienta.2022.111695. - DOI
-
- Li Y.Y., Guo L.L., Wang Z.Y., Zhao D.H., Guo D.L., Carlson J.E., Yin W.L., Hou X.G. Genome-wide association study of 23 flowering phenology traits and 4 floral agronomic traits in tree peony (Paeonia section Moutan DC.) reveals five genes known to regulate flowering time. Hortic. Res. 2023;10:uhac263. doi: 10.1093/hr/uhac263. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

