DREB1 and DREB2 Genes in Garlic (Allium sativum L.): Genome-Wide Identification, Characterization, and Stress Response
- PMID: 37447098
- PMCID: PMC10347091
- DOI: 10.3390/plants12132538
DREB1 and DREB2 Genes in Garlic (Allium sativum L.): Genome-Wide Identification, Characterization, and Stress Response
Abstract
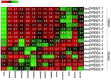
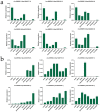
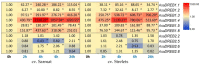
Dehydration-responsive element-binding (DREB) transcription factors (TFs) of the A1 and A2 subfamilies involved in plant stress responses have not yet been reported in Allium species. In this study, we used bioinformatics and comparative transcriptomics to identify and characterize DREB A1 and A2 genes redundant in garlic (Allium sativum L.) and analyze their expression in A. sativum cultivars differing in the sensitivity to cold and Fusarium infection. Eight A1 (AsaDREB1.1-1.8) and eight A2 (AsaDREB2.1-2.8) genes were identified. AsaDREB1.1-1.8 genes located in tandem on chromosome 1 had similar expression patterns, suggesting functional redundancy. AsaDREB2.1-2.8 were scattered on different chromosomes and had organ- and genotype-specific expressions. AsaDREB1 and AsaDREB2 promoters contained 7 and 9 hormone- and stress-responsive cis-regulatory elements, respectively, and 13 sites associated with TF binding and plant development. In both Fusarium-resistant and -sensitive cultivars, fungal infection upregulated the AsaDREB1.1-1.5, 1.8, 2.2, 2.6, and 2.8 genes and downregulated AsaDREB2.5, but the magnitude of response depended on the infection susceptibility of the cultivar. Cold exposure strongly upregulated the AsaDREB1 genes, but downregulated most AsaDREB2 genes. Our results provide the foundation for further functional analysis of the DREB TFs in Allium crops and could contribute to the breeding of stress-tolerant varieties.
Keywords: Allium sativum L.; DREB proteins; Fusarium; cold stress; gene expression; gene structure.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Thaumatin-like Protein (TLP) Genes in Garlic (Allium sativum L.): Genome-Wide Identification, Characterization, and Expression in Response to Fusarium proliferatum Infection.Plants (Basel). 2022 Mar 11;11(6):748. doi: 10.3390/plants11060748. Plants (Basel). 2022. PMID: 35336630 Free PMC article.
-
Identification of Chalcone Synthase Genes from Garlic (Allium sativum L.) and Their Expression Levels in Response to Stress Factors.Acta Naturae. 2025 Apr-Jun;17(2):41-51. doi: 10.32607/actanaturae.27639. Acta Naturae. 2025. PMID: 40771850 Free PMC article.
-
Pathogenesis-Related Genes of PR1, PR2, PR4, and PR5 Families Are Involved in the Response to Fusarium Infection in Garlic (Allium sativum L.).Int J Mol Sci. 2021 Jun 22;22(13):6688. doi: 10.3390/ijms22136688. Int J Mol Sci. 2021. PMID: 34206508 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
Cited by
-
Combined Bulked Segregant Analysis-Sequencing and Transcriptome Analysis to Identify Candidate Genes Associated with Cold Stress in Brassica napus L.Int J Mol Sci. 2025 Jan 28;26(3):1148. doi: 10.3390/ijms26031148. Int J Mol Sci. 2025. PMID: 39940915 Free PMC article.
-
Integrated genome-wide association and transcriptomic studies reveal genetic architecture of bulb storability of plentiful garlic germplasm resources.Hortic Res. 2024 Sep 16;11(12):uhae260. doi: 10.1093/hr/uhae260. eCollection 2024 Dec. Hortic Res. 2024. PMID: 39664692 Free PMC article.
References
-
- El-Saber Batiha G., Magdy Beshbishy A., Wasef L.G., Elewa Y.H.A., Al-Sagan A.A., Abd El-Hack M.E., Taha A.E., Abd-Elhakim Y.M., Prasad Devkota H. Chemical constituents and pharmacological activities of garlic (Allium sativum L.): A Review. Nutrients. 2020;12:872. doi: 10.3390/nu12030872. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

