Genome-Wide Association Study of Xian Rice Grain Shape and Weight in Different Environments
- PMID: 37447110
- PMCID: PMC10347298
- DOI: 10.3390/plants12132549
Genome-Wide Association Study of Xian Rice Grain Shape and Weight in Different Environments
Abstract
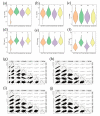
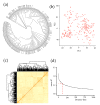
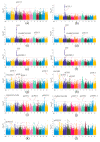
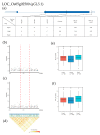
Drought is one of the key environmental factors affecting the growth and yield potential of rice. Grain shape, on the other hand, is an important factor determining the appearance, quality, and yield of rice grains. Here, we re-sequenced 275 Xian accessions and then conducted a genome-wide association study (GWAS) on six agronomic traits with the 404,411 single nucleotide polymorphisms (SNPs) derived by the best linear unbiased prediction (BLUP) for each trait. Under two years of drought stress (DS) and normal water (NW) treatments, a total of 16 QTLs associated with rice grain shape and grain weight were detected on chromosomes 1, 2, 3, 4, 5, 7, 8, 11, and 12. In addition, these QTLs were analyzed by haplotype analysis and functional annotation, and one clone (GSN1) and five new candidate genes were identified in the candidate interval. The findings provide important genetic information for the molecular improvement of grain shape and weight in rice.
Keywords: GWAS; QTLs; grain shape; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

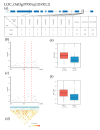

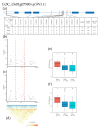
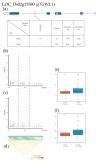
References
-
- Zahra N., Hafeez M.B., Nawaz A., Farooq M. Rice production systems and grain quality. J. Cereal Sci. 2022;105:103463. doi: 10.1016/j.jcs.2022.103463. - DOI
-
- Calingacion M., Laborte A., Nelson A., Resurreccion A., Concepcion J.C., Daygon V.D., Mumm R., Reinke R., Dipti S., Bassinello P.Z., et al. Diversity of Global Rice Markets and the Science Required for Consumer-Targeted Rice Breeding. PLoS ONE. 2014;9:12. doi: 10.1371/journal.pone.0085106. - DOI - PMC - PubMed
-
- Russinga A.M. Correlation Studies on Yield and Yield Contributing Traits in Rice (Oryza sativa L.) Ind. J. Pure App. Biosci. 2020;8:531–538. doi: 10.18782/2582-2845.8334. - DOI
Grants and funding
- 2022AH040126/The Improved Varieties Joint Research (Rice) Project of Anhui Province (the 14th fifive-year plan), the Scientific Research Plan Major Projects of Anhui Province
- 2021d06050002/The Science and Technology Major Project of Anhui Province
- U21A20214,31971927/The National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

