Dynamic structure of T4 gene 32 protein filaments facilitates rapid noncooperative protein dissociation
- PMID: 37449435
- PMCID: PMC10484735
- DOI: 10.1093/nar/gkad595
Dynamic structure of T4 gene 32 protein filaments facilitates rapid noncooperative protein dissociation
Erratum in
-
Correction to 'Dynamic structure of T4 gene 32 protein filaments facilitates rapid noncooperative protein dissociation'.Nucleic Acids Res. 2023 Oct 27;51(19):10811. doi: 10.1093/nar/gkad780. Nucleic Acids Res. 2023. PMID: 37738142 Free PMC article. No abstract available.
Abstract
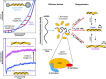
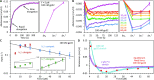
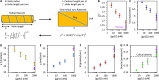
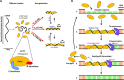
Bacteriophage T4 gene 32 protein (gp32) is a model single-stranded DNA (ssDNA) binding protein, essential for DNA replication. gp32 forms cooperative filaments on ssDNA through interprotein interactions between its core and N-terminus. However, detailed understanding of gp32 filament structure and organization remains incomplete, particularly for longer, biologically-relevant DNA lengths. Moreover, it is unclear how these tightly-bound filaments dissociate from ssDNA during complementary strand synthesis. We use optical tweezers and atomic force microscopy to probe the structure and binding dynamics of gp32 on long (∼8 knt) ssDNA substrates. We find that cooperative binding of gp32 rigidifies ssDNA while also reducing its contour length, consistent with the ssDNA helically winding around the gp32 filament. While measured rates of gp32 binding and dissociation indicate nM binding affinity, at ∼1000-fold higher protein concentrations gp32 continues to bind into and restructure the gp32-ssDNA filament, leading to an increase in its helical pitch and elongation of the substrate. Furthermore, the oversaturated gp32-ssDNA filament becomes progressively unwound and unstable as observed by the appearance of a rapid, noncooperative protein dissociation phase not seen at lower complex saturation, suggesting a possible mechanism for prompt removal of gp32 from the overcrowded ssDNA in front of the polymerase during replication.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







Similar articles
-
C-terminal Domain of T4 gene 32 Protein Enables Rapid Filament Reorganization and Dissociation.J Mol Biol. 2024 May 1;436(9):168544. doi: 10.1016/j.jmb.2024.168544. Epub 2024 Mar 18. J Mol Biol. 2024. PMID: 38508303
-
Assembly and dynamics of Gp59-Gp32-single-stranded DNA (ssDNA), a DNA helicase loading complex required for recombination-dependent replication in bacteriophage T4.J Biol Chem. 2012 Jun 1;287(23):19070-81. doi: 10.1074/jbc.M112.343830. Epub 2012 Apr 12. J Biol Chem. 2012. PMID: 22500043 Free PMC article.
-
Helicase assembly protein Gp59 of bacteriophage T4: fluorescence anisotropy and sedimentation studies of complexes formed with derivatives of Gp32, the phage ssDNA binding protein.Biochemistry. 2001 Jun 26;40(25):7651-61. doi: 10.1021/bi010116n. Biochemistry. 2001. PMID: 11412119
-
Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions.J Biol Chem. 2006 Sep 8;281(36):26308-19. doi: 10.1074/jbc.M604349200. Epub 2006 Jul 7. J Biol Chem. 2006. PMID: 16829679
-
Optical tweezers experiments resolve distinct modes of DNA-protein binding.Biopolymers. 2009 Apr;91(4):265-82. doi: 10.1002/bip.21123. Biopolymers. 2009. PMID: 19173290 Free PMC article. Review.
Cited by
-
The mutation R107Q alters mtSSB ssDNA compaction ability and binding dynamics.Nucleic Acids Res. 2024 Jun 10;52(10):5912-5927. doi: 10.1093/nar/gkae354. Nucleic Acids Res. 2024. PMID: 38742632 Free PMC article.
-
Structural basis for cooperative ssDNA binding by bacteriophage protein filament P12.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf132. doi: 10.1093/nar/gkaf132. Nucleic Acids Res. 2025. PMID: 40052821 Free PMC article.
-
DNA damage alters binding conformations of E. coli single-stranded DNA-binding protein.Biophys J. 2023 Oct 3;122(19):3950-3958. doi: 10.1016/j.bpj.2023.08.018. Epub 2023 Aug 24. Biophys J. 2023. PMID: 37632138 Free PMC article.
-
HIV-1 uncoating requires long double-stranded reverse transcription products.Sci Adv. 2024 Apr 26;10(17):eadn7033. doi: 10.1126/sciadv.adn7033. Epub 2024 Apr 24. Sci Adv. 2024. PMID: 38657061 Free PMC article.
-
L1-ORF1p nucleoprotein can rapidly assume distinct conformations and simultaneously bind more than one nucleic acid.Nucleic Acids Res. 2024 Dec 11;52(22):14013-14029. doi: 10.1093/nar/gkae1141. Nucleic Acids Res. 2024. PMID: 39565204 Free PMC article.
References
-
- Alberts B.M. Prokaryotic DNA replication mechanisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1987; 317:395–420. - PubMed
-
- Johnson A., O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005; 74:283–315. - PubMed
-
- Nossal N. Molecular biology of bacteriophage T4. Am. Soc. Microbiol. 1994; 43–53.
-
- Alberts B.M., Frey L.. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970; 227:1313–1318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

