Quantitative proteomic and phosphoproteomic analyses of the hippocampus reveal the involvement of NMDAR1 signaling in repetitive mild traumatic brain injury
- PMID: 37449635
- PMCID: PMC10358661
- DOI: 10.4103/1673-5374.374654
Quantitative proteomic and phosphoproteomic analyses of the hippocampus reveal the involvement of NMDAR1 signaling in repetitive mild traumatic brain injury
Abstract
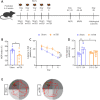
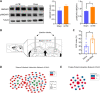
The cumulative damage caused by repetitive mild traumatic brain injury can cause long-term neurodegeneration leading to cognitive impairment. This cognitive impairment is thought to result specifically from damage to the hippocampus. In this study, we detected cognitive impairment in mice 6 weeks after repetitive mild traumatic brain injury using the novel object recognition test and the Morris water maze test. Immunofluorescence staining showed that p-tau expression was increased in the hippocampus after repetitive mild traumatic brain injury. Golgi staining showed a significant decrease in the total density of neuronal dendritic spines in the hippocampus, as well as in the density of mature dendritic spines. To investigate the specific molecular mechanisms underlying cognitive impairment due to hippocampal damage, we performed proteomic and phosphoproteomic analyses of the hippocampus with and without repetitive mild traumatic brain injury. The differentially expressed proteins were mainly enriched in inflammation, immunity, and coagulation, suggesting that non-neuronal cells are involved in the pathological changes that occur in the hippocampus in the chronic stage after repetitive mild traumatic brain injury. In contrast, differentially expressed phosphorylated proteins were mainly enriched in pathways related to neuronal function and structure, which is more consistent with neurodegeneration. We identified N-methyl-D-aspartate receptor 1 as a hub molecule involved in the response to repetitive mild traumatic brain injury , and western blotting showed that, while N-methyl-D-aspartate receptor 1 expression was not altered in the hippocampus after repetitive mild traumatic brain injury, its phosphorylation level was significantly increased, which is consistent with the omics results. Administration of GRP78608, an N-methyl-D-aspartate receptor 1 antagonist, to the hippocampus markedly improved repetitive mild traumatic brain injury-induced cognitive impairment. In conclusion, our findings suggest that N-methyl-D-aspartate receptor 1 signaling in the hippocampus is involved in cognitive impairment in the chronic stage after repetitive mild traumatic brain injury and may be a potential target for intervention and treatment.
Keywords: Grin1; N-methyl-D-aspartate; N-methyl-D-aspartate receptor 1; cognitive impairment; hippocampus; learning; memory; phosphoproteomic; proteomic; repetitive mild traumatic brain injury (rmTBI); secondary injury.
Conflict of interest statement
None
Figures




References
-
- Apweiler R, Biswas M, Fleischmann W, Kanapin A, Karavidopoulou Y, Kersey P, Kriventseva EV, Mittard V, Mulder N, Phan I, Zdobnov E. Proteome analysis database:online application of interpro and clustr for the functional classification of proteins in whole genomes. Nucleic Acids Res. 2001;29:44–48. - PMC - PubMed
-
- Ayubcha C, Revheim ME, Newberg A, Moghbel M, Rojulpote C, Werner TJ, Alavi A. A critical review of radiotracers in the positron emission tomography imaging of traumatic brain injury:fdg, tau, and amyloid imaging in mild traumatic brain injury and chronic traumatic encephalopathy. Eur J Nucl Med Mol Imaging. 2021;48:623–641. - PubMed
LinkOut - more resources
Full Text Sources

