ENIGMA CHEK2gether Project: A Comprehensive Study Identifies Functionally Impaired CHEK2 Germline Missense Variants Associated with Increased Breast Cancer Risk
- PMID: 37449874
- PMCID: PMC10425727
- DOI: 10.1158/1078-0432.CCR-23-0212
ENIGMA CHEK2gether Project: A Comprehensive Study Identifies Functionally Impaired CHEK2 Germline Missense Variants Associated with Increased Breast Cancer Risk
Abstract
Purpose: Germline pathogenic variants in CHEK2 confer moderately elevated breast cancer risk (odds ratio, OR ∼ 2.5), qualifying carriers for enhanced breast cancer screening. Besides pathogenic variants, dozens of missense CHEK2 variants of uncertain significance (VUS) have been identified, hampering the clinical utility of germline genetic testing (GGT).
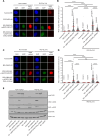
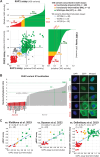
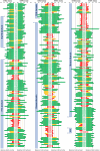
Experimental design: We collected 460 CHEK2 missense VUS identified by the ENIGMA consortium in 15 countries. Their functional characterization was performed using CHEK2-complementation assays quantifying KAP1 phosphorylation and CHK2 autophosphorylation in human RPE1-CHEK2-knockout cells. Concordant results in both functional assays were used to categorize CHEK2 VUS from 12 ENIGMA case-control datasets, including 73,048 female patients with breast cancer and 88,658 ethnicity-matched controls.
Results: A total of 430/460 VUS were successfully analyzed, of which 340 (79.1%) were concordant in both functional assays and categorized as functionally impaired (N = 102), functionally intermediate (N = 12), or functionally wild-type (WT)-like (N = 226). We then examined their association with breast cancer risk in the case-control analysis. The OR and 95% CI (confidence intervals) for carriers of functionally impaired, intermediate, and WT-like variants were 2.83 (95% CI, 2.35-3.41), 1.57 (95% CI, 1.41-1.75), and 1.19 (95% CI, 1.08-1.31), respectively. The meta-analysis of population-specific datasets showed similar results.
Conclusions: We determined the functional consequences for the majority of CHEK2 missense VUS found in patients with breast cancer (3,660/4,436; 82.5%). Carriers of functionally impaired missense variants accounted for 0.5% of patients with breast cancer and were associated with a moderate risk similar to that of truncating CHEK2 variants. In contrast, 2.2% of all patients with breast cancer carried functionally wild-type/intermediate missense variants with no clinically relevant breast cancer risk in heterozygous carriers.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 1999;286:2528–31. - PubMed
-
- Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol 2008;26:542–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

