Endocrine Therapy Synergizes with SMAC Mimetics to Potentiate Antigen Presentation and Tumor Regression in Hormone Receptor-Positive Breast Cancer
- PMID: 37450351
- PMCID: PMC10543960
- DOI: 10.1158/0008-5472.CAN-23-1711
Endocrine Therapy Synergizes with SMAC Mimetics to Potentiate Antigen Presentation and Tumor Regression in Hormone Receptor-Positive Breast Cancer
Abstract
Immunotherapies have yet to demonstrate significant efficacy in the treatment of hormone receptor-positive (HR+) breast cancer. Given that endocrine therapy (ET) is the primary approach for treating HR+ breast cancer, we investigated the effects of ET on the tumor immune microenvironment (TME) in HR+ breast cancer. Spatial proteomics of primary HR+ breast cancer samples obtained at baseline and after ET from patients enrolled in a neoadjuvant clinical trial (NCT02764541) indicated that ET upregulated β2-microglobulin and influenced the TME in a manner that promotes enhanced immunogenicity. To gain a deeper understanding of the underlying mechanisms, the intrinsic effects of ET on cancer cells were explored, which revealed that ET plays a crucial role in facilitating the chromatin binding of RelA, a key component of the NF-κB complex. Consequently, heightened NF-κB signaling enhanced the response to interferon-gamma, leading to the upregulation of β2-microglobulin and other antigen presentation-related genes. Further, modulation of NF-κB signaling using a SMAC mimetic in conjunction with ET augmented T-cell migration and enhanced MHC-I-specific T-cell-mediated cytotoxicity. Remarkably, the combination of ET and SMAC mimetics, which also blocks prosurvival effects of NF-κB signaling through the degradation of inhibitors of apoptosis proteins, elicited tumor regression through cell autonomous mechanisms, providing additional support for their combined use in HR+ breast cancer.
Significance: Adding SMAC mimetics to endocrine therapy enhances tumor regression in a cell autonomous manner while increasing tumor immunogenicity, indicating that this combination could be an effective treatment for HR+ patients with breast cancer.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures

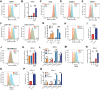
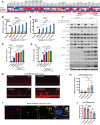
![Figure 2. Endocrine treatment shapes tumor immune microenvironment in primary hormone receptor–positive breast cancer. A–F, Digital spatial profiling proteomic levels (log2 expression levels) of immune cell surface markers ranking from highest to lowest expression. A, Baseline levels within the immune regions in tumors from patients who received endocrine therapy [F(11, 312) = 57.1, P < 2e−16, one-way ANOVA]. B, Protein expression levels within the immune regions after 2 weeks of endocrine treatment [F(11, 276) = 41.7, P < 2e−16, one-way ANOVA]. C, Protein expression levels within the immune regions after 24 weeks of endocrine treatment at the time of surgery [F(11, 1212) = 305.2, P < 2e−16, one-way ANOVA]. D, Baseline levels within the invasive epithelial cell regions in tumors from patients who received endocrine therapy [F (11, 732) = 126.3, P < 2e−16, one-way ANOVA]. E, Protein expression levels within the invasive epithelial regions after 2 weeks of endocrine treatment (P < 2e−16, one-way ANOVA). F, Protein expression levels within the invasive epithelial cell regions after 24 weeks of endocrine treatment at the time of surgery (P < 2e−16, one-way ANOVA). Boxplots show median, 25th, and 75th percentiles as boxes, the minimum of the 75th percentile + 1.5 × IQR, and the maximum observation as the upper whisker and the maximum of the 25th percentile −1.5 × IQR and the minimum observation as the lower whisker. G, Trajectory plot of TIL fractions between baseline and 2 weeks. P-val, P value (paired Wilcoxon signed-rank test with continuity correction). H, Trajectory plot of TIL fractions between 2 weeks and surgery for all patients given endocrine treatment that have TIL observations at all three time points. Each trajectory corresponds to a single patient. P-val, P value (paired Wilcoxon signed-rank test with continuity correction). I, GSVA of the T-cell accumulation gene set in primary ER+ breast cancer biopsies from pre- and post-neoadjuvant AI treatment. J, Enrichment plot of the top-ranked gene set (estrogen response) enriched in the ESR1 highest (fourth quartile) versus ESR1 lowest (first quartile) ER–positive breast cancer samples from the TCGA cohort. K–M, GSVA of RNA-seq from the ER–positive breast cancer samples from TCGA divided into quartiles based on ESR1 mRNA levels testing the enrichment score (y-axis) for signatures of immune-checkpoint blockade (ICB) resistance (K), T-regulatory (Treg) accumulation (L), and cytotoxic T-cell accumulation (M). Comparison between the quartiles was done with a t test. N, number of patients included in the corresponding analysis.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/510f/10543960/d396f215d702/3284fig2.gif)



References
-
- Korde LA, Somerfield MR, Hershman DL; Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer Guideline Expert Panel. Use of immune checkpoint inhibitor pembrolizumab in the treatment of high-risk, early-stage triple-negative breast cancer: ASCO guideline rapid recommendation update. J Clin Oncol 2022;40:1696–8. - PubMed
-
- Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020;396:1817–28. - PubMed
-
- Rugo HS, Delord JP, Im SA, Ott PA, Piha-Paul SA, Bedard PL, et al. Safety and antitumor activity of pembrolizumab in patients with estrogen receptor-positive/human epidermal growth factor receptor 2-negative advanced breast cancer. Clin Cancer Res 2018;24:2804–11. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

