Dynamics-informed deconvolutional neural networks for super-resolution identification of regime changes in epidemiological time series
- PMID: 37450598
- PMCID: PMC10348669
- DOI: 10.1126/sciadv.adf0673
Dynamics-informed deconvolutional neural networks for super-resolution identification of regime changes in epidemiological time series
Abstract
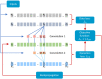
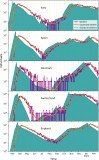
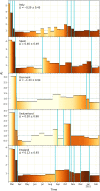
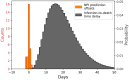
The ability to infer the timing and amplitude of perturbations in epidemiological systems from their stochastically spread low-resolution outcomes is crucial for multiple applications. However, the general problem of connecting epidemiological curves with the underlying incidence lacks the highly effective methodology present in other inverse problems, such as super-resolution and dehazing from computer vision. Here, we develop an unsupervised physics-informed convolutional neural network approach in reverse to connect death records with incidence that allows the identification of regime changes at single-day resolution. Applied to COVID-19 data with proper regularization and model-selection criteria, the approach can identify the implementation and removal of lockdowns and other nonpharmaceutical interventions (NPIs) with 0.93-day accuracy over the time span of a year.
Figures

References
-
- R. Szeliski, Computer Vision: Algorithms and Applications. (Springer-Verlag, ed. 2, 2022).
-
- C. Payer, D. Štern, H. Bischof, M. Urschler, Regressing heatmaps for multiple landmark localization using CNNs, in Medical Image Computing and Computer-Assisted Intervention (2016), vol. 9901, pp. 230–238.
-
- Li B., Ren W., Fu D., Tao D., Feng D., Zeng W., Wang Z., Benchmarking single-image dehazing and beyond. IEEE Trans. Image Process. 28, 492–505 (2019). - PubMed
-
- Biemond J., Lagendijk R. L., Mersereau R. M., Iterative methods for image deblurring. Proc. IEEE 78, 856–883 (1990).
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

