Hypoxic niches attract and sequester tumor-associated macrophages and cytotoxic T cells and reprogram them for immunosuppression
- PMID: 37451265
- PMCID: PMC10527169
- DOI: 10.1016/j.immuni.2023.06.017
Hypoxic niches attract and sequester tumor-associated macrophages and cytotoxic T cells and reprogram them for immunosuppression
Abstract
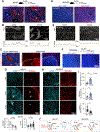
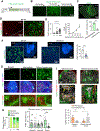
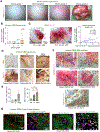
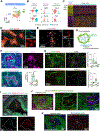
Glioblastoma (GBM), a highly lethal brain cancer, is notorious for immunosuppression, but the mechanisms remain unclear. Here, we documented a temporospatial patterning of tumor-associated myeloid cells (TAMs) corresponding to vascular changes during GBM progression. As tumor vessels transitioned from the initial dense regular network to later scant and engorged vasculature, TAMs shifted away from perivascular regions and trafficked to vascular-poor areas. This process was heavily influenced by the immunocompetence state of the host. Utilizing a sensitive fluorescent UnaG reporter to track tumor hypoxia, coupled with single-cell transcriptomics, we revealed that hypoxic niches attracted and sequestered TAMs and cytotoxic T lymphocytes (CTLs), where they were reprogrammed toward an immunosuppressive state. Mechanistically, we identified chemokine CCL8 and cytokine IL-1β as two hypoxic-niche factors critical for TAM trafficking and co-evolution of hypoxic zones into pseudopalisading patterns. Therefore, perturbation of TAM patterning in hypoxic zones may improve tumor control.
Keywords: CCL8; CTLs; GBM; IL-1β; TAM; cytotoxic T lymphocytes; immune landscape; immunosuppression; tumor hypoxia; tumor vasculature; tumor-associated microglia/macrophages.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Comment in
-
Take my breath away: TAM-ing anti-cancer immunity in hypoxic niches.Immunity. 2023 Aug 8;56(8):1704-1706. doi: 10.1016/j.immuni.2023.07.010. Immunity. 2023. PMID: 37557077
References
-
- Klemm F, Maas RR, Bowman RL, Kornete M, Soukup K, Nassiri S, Brouland JP, Iacobuzio-Donahue CA, Brennan C, Tabar V, et al. (2020). Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 181, 1643–1660.e1617. 10.1016/j.cell.2020.05.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

