Negative regulation of MurZ and MurA underlies the essentiality of GpsB- and StkP-mediated protein phosphorylation in Streptococcus pneumoniae D39
- PMID: 37452010
- PMCID: PMC10530524
- DOI: 10.1111/mmi.15122
Negative regulation of MurZ and MurA underlies the essentiality of GpsB- and StkP-mediated protein phosphorylation in Streptococcus pneumoniae D39
Abstract
GpsB links peptidoglycan synthases to other proteins that determine the shape of the respiratory pathogen Streptococcus pneumoniae (pneumococcus; Spn) and other low-GC Gram-positive bacteria. GpsB is also required for phosphorylation of proteins by the essential StkP(Spn) Ser/Thr protein kinase. Here we report three classes of frequently arising chromosomal duplications (≈21-176 genes) containing murZ (MurZ-family homolog of MurA) or murA that suppress ΔgpsB or ΔstkP. These duplications arose from three different repeated sequences and demonstrate the facility of pneumococcus to modulate gene dosage of numerous genes. Overproduction of MurZ or MurA alone or overproduction of MurZ caused by ΔkhpAB mutations suppressed ΔgpsB or ΔstkP phenotypes to varying extents. ΔgpsB and ΔstkP were also suppressed by MurZ amino-acid changes distant from the active site, including one in commonly studied laboratory strains, and by truncation or deletion of the homolog of IreB(ReoM). Unlike in other Gram-positive bacteria, MurZ is predominant to MurA in pneumococcal cells. However, ΔgpsB and ΔstkP were not suppressed by ΔclpCP, which did not alter MurZ or MurA amounts. These results support a model in which regulation of MurZ and MurA activity, likely by IreB(Spn), is the only essential requirement for StkP-mediated protein phosphorylation in exponentially growing D39 pneumococcal cells.
Keywords: GpsB peptidoglycan regulator; KhpA/B RNA binding protein; StkP protein kinase; gene duplication and amplification; peptidoglycan precursor synthesis.
© 2023 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interests.
Figures

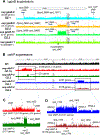


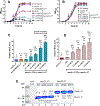





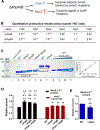

Update of
-
Chromosomal Duplications of MurZ (MurA2) or MurA (MurA1), Amino Acid Substitutions in MurZ (MurA2), and Absence of KhpAB Obviate the Requirement for Protein Phosphorylation in Streptococcus pneumoniae D39.bioRxiv [Preprint]. 2023 Mar 27:2023.03.26.534294. doi: 10.1101/2023.03.26.534294. bioRxiv. 2023. Update in: Mol Microbiol. 2023 Sep;120(3):351-383. doi: 10.1111/mmi.15122. PMID: 37034771 Free PMC article. Updated. Preprint.
Similar articles
-
Chromosomal Duplications of MurZ (MurA2) or MurA (MurA1), Amino Acid Substitutions in MurZ (MurA2), and Absence of KhpAB Obviate the Requirement for Protein Phosphorylation in Streptococcus pneumoniae D39.bioRxiv [Preprint]. 2023 Mar 27:2023.03.26.534294. doi: 10.1101/2023.03.26.534294. bioRxiv. 2023. Update in: Mol Microbiol. 2023 Sep;120(3):351-383. doi: 10.1111/mmi.15122. PMID: 37034771 Free PMC article. Updated. Preprint.
-
Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39.Mol Microbiol. 2017 Mar;103(6):931-957. doi: 10.1111/mmi.13613. Epub 2017 Feb 7. Mol Microbiol. 2017. PMID: 28010038 Free PMC article.
-
Suppressor Mutations Linking gpsB with the First Committed Step of Peptidoglycan Biosynthesis in Listeria monocytogenes.J Bacteriol. 2016 Dec 13;199(1):e00393-16. doi: 10.1128/JB.00393-16. Print 2017 Jan 1. J Bacteriol. 2016. PMID: 27795316 Free PMC article.
-
Cell division of Streptococcus pneumoniae: think positive!Curr Opin Microbiol. 2016 Dec;34:18-23. doi: 10.1016/j.mib.2016.07.014. Epub 2016 Aug 4. Curr Opin Microbiol. 2016. PMID: 27497051 Review.
-
From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division?Environ Microbiol. 2013 Dec;15(12):3133-57. doi: 10.1111/1462-2920.12189. Epub 2013 Jul 15. Environ Microbiol. 2013. PMID: 23848140 Review.
Cited by
-
GpsB Coordinates StkP Signaling as a PASTA Kinase Adaptor in Streptococcus pneumoniae Cell Division.J Mol Biol. 2024 Nov 15;436(22):168797. doi: 10.1016/j.jmb.2024.168797. Epub 2024 Sep 19. J Mol Biol. 2024. PMID: 39303764
-
Secrets of getting started: Regulation of the first committed step of peptidoglycan synthesis by protein phosphorylation in Enterococcus and other Gram-positive bacteria.Mol Microbiol. 2023 Dec;120(6):805-810. doi: 10.1111/mmi.15204. Epub 2023 Nov 27. Mol Microbiol. 2023. PMID: 38012814 Free PMC article.
-
Biological puzzles solved by using Streptococcus pneumoniae: a historical review of the pneumococcal studies that have impacted medicine and shaped molecular bacteriology.J Bacteriol. 2024 Jun 20;206(6):e0005924. doi: 10.1128/jb.00059-24. Epub 2024 May 29. J Bacteriol. 2024. PMID: 38809015 Free PMC article. Review.
-
Elongasome core proteins and class A PBP1a display zonal, processive movement at the midcell of Streptococcus pneumoniae.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2401831121. doi: 10.1073/pnas.2401831121. Epub 2024 Jun 14. Proc Natl Acad Sci U S A. 2024. PMID: 38875147 Free PMC article.
-
Alternate routes to mnm5s2U synthesis in Gram-positive bacteria.J Bacteriol. 2024 Apr 18;206(4):e0045223. doi: 10.1128/jb.00452-23. Epub 2024 Mar 29. J Bacteriol. 2024. PMID: 38551342 Free PMC article.
References
-
- Aguilan JT, Kulej K, and Sidoli S (2020) Guide for protein fold change and p-value calculation for non-experts in proteomics. Mol Omics 16: 573–582. - PubMed
-
- Alonso CA, Dominguez C, Heras J, Mata E, Pascual V, Torres C, and Zarazaga M (2017) Antibiogramj: A tool for analysing images from disk diffusion tests. Comput Methods Programs Biomed 143: 159–169. - PubMed
-
- Blake KL, O’Neill AJ, Mengin-Lecreulx D, Henderson PJ, Bostock JM, Dunsmore CJ, Simmons KJ, Fishwick CW, Leeds JA, and Chopra I (2009) The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol Microbiol 72: 335–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

