Distinct evolution of type I glutamine synthetase in Plasmodium and its species-specific requirement
- PMID: 37452051
- PMCID: PMC10349072
- DOI: 10.1038/s41467-023-39670-4
Distinct evolution of type I glutamine synthetase in Plasmodium and its species-specific requirement
Abstract
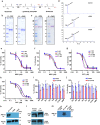
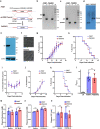
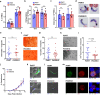
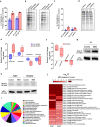
Malaria parasite lacks canonical pathways for amino acid biosynthesis and depends primarily on hemoglobin degradation and extracellular resources for amino acids. Interestingly, a putative gene for glutamine synthetase (GS) is retained despite glutamine being an abundant amino acid in human and mosquito hosts. Here we show Plasmodium GS has evolved as a unique type I enzyme with distinct structural and regulatory properties to adapt to the asexual niche. Methionine sulfoximine (MSO) and phosphinothricin (PPT) inhibit parasite GS activity. GS is localized to the parasite cytosol and abundantly expressed in all the life cycle stages. Parasite GS displays species-specific requirement in Plasmodium falciparum (Pf) having asparagine-rich proteome. Targeting PfGS affects asparagine levels and inhibits protein synthesis through eIF2α phosphorylation leading to parasite death. Exposure of artemisinin-resistant Pf parasites to MSO and PPT inhibits the emergence of viable parasites upon artemisinin treatment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

