Efferocytosis by bone marrow mesenchymal stromal cells disrupts osteoblastic differentiation via mitochondrial remodeling
- PMID: 37452070
- PMCID: PMC10349065
- DOI: 10.1038/s41419-023-05931-9
Efferocytosis by bone marrow mesenchymal stromal cells disrupts osteoblastic differentiation via mitochondrial remodeling
Abstract
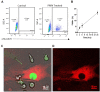
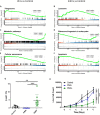
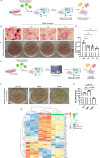
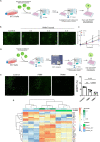
The efficient clearance of dead and dying cells, efferocytosis, is critical to maintain tissue homeostasis. In the bone marrow microenvironment (BMME), this role is primarily fulfilled by professional bone marrow macrophages, but recent work has shown that mesenchymal stromal cells (MSCs) act as a non-professional phagocyte within the BMME. However, little is known about the mechanism and impact of efferocytosis on MSCs and on their function. To investigate, we performed flow cytometric analysis of neutrophil uptake by ST2 cells, a murine bone marrow-derived stromal cell line, and in murine primary bone marrow-derived stromal cells. Transcriptional analysis showed that MSCs possess the necessary receptors and internal processing machinery to conduct efferocytosis, with Axl and Tyro3 serving as the main receptors, while MerTK was not expressed. Moreover, the expression of these receptors was modulated by efferocytic behavior, regardless of apoptotic target. MSCs derived from human bone marrow also demonstrated efferocytic behavior, showing that MSC efferocytosis is conserved. In all MSCs, efferocytosis impaired osteoblastic differentiation. Transcriptional analysis and functional assays identified downregulation in MSC mitochondrial function upon efferocytosis. Experimentally, efferocytosis induced mitochondrial fission in MSCs. Pharmacologic inhibition of mitochondrial fission in MSCs not only decreased efferocytic activity but also rescued osteoblastic differentiation, demonstrating that efferocytosis-mediated mitochondrial remodeling plays a critical role in regulating MSC differentiation. This work describes a novel function of MSCs as non-professional phagocytes within the BMME and demonstrates that efferocytosis by MSCs plays a key role in directing mitochondrial remodeling and MSC differentiation. Efferocytosis by MSCs may therefore be a novel mechanism of dysfunction and senescence. Since our data in human MSCs show that MSC efferocytosis is conserved, the consequences of MSC efferocytosis may impact the behavior of these cells in the human skeleton, including bone marrow remodeling and bone loss in the setting of aging, cancer and other diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

