Plastome evolution and phylogenomic insights into the evolution of Lysimachia (Primulaceae: Myrsinoideae)
- PMID: 37452336
- PMCID: PMC10347800
- DOI: 10.1186/s12870-023-04363-z
Plastome evolution and phylogenomic insights into the evolution of Lysimachia (Primulaceae: Myrsinoideae)
Abstract
Background: Lysimachia L., the second largest genus within the subfamily Myrsinoideae of Primulaceae, comprises approximately 250 species worldwide. China is the species diversity center of Lysimachia, containing approximately 150 species. Despite advances in the backbone phylogeny of Lysimachia, species-level relationships remain poorly understood due to limited genomic information. This study analyzed 50 complete plastomes for 46 Lysimachia species. We aimed to identify the plastome structure features and hypervariable loci of Lysimachia. Additionally, the phylogenetic relationships and phylogenetic conflict signals in Lysimachia were examined.
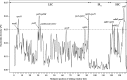
Results: These fifty plastomes within Lysimachia had the typical quadripartite structure, with lengths varying from 152,691 to 155,784 bp. Plastome size was positively correlated with IR and intron length. Thirteen highly variable regions in Lysimachia plastomes were identified. Additionally, ndhB, petB and ycf2 were found to be under positive selection. Plastid ML trees and species tree strongly supported that L. maritima as sister to subg. Palladia + subg. Lysimachia (Christinae clade), while the nrDNA ML tree clearly placed L. maritima and subg. Palladia as a sister group.
Conclusions: The structures of these plastomes of Lysimachia were generally conserved, but potential plastid markers and signatures of positive selection were detected. These genomic data provided new insights into the interspecific relationships of Lysimachia, including the cytonuclear discordance of the position of L. maritima, which may be the result of ghost introgression in the past. Our findings have established a basis for further exploration of the taxonomy, phylogeny and evolutionary history within Lysimachia.
Keywords: Conflict signature; Lysimachia; Phylogeny; Plastid genome; Primulaceae.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Hu CM, Kelso S. Primulaceae. In: Flora of China. Edited by Wu CY, Raven PH. vol. 15. St Louis, USA/Beijing, China: Missouri Botanical Garden Press/Science Press; 1996: 39–189.
-
- Anderberg AA, Manns U, Källersjö M. Phylogeny and floral evolution of the Lysimachieae (Ericales, Myrsinaceae): evidence from ndhF sequence data. Willdenowia. 2007;37(2):407–421. doi: 10.3372/wi.37.37202. - DOI
-
- Ståhl B, Anderberg AA. Myrsinaceae. In: Flowering plants · dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales. Edited by Kubitzki K. Berlin, Heidelberg: Springer Berlin Heidelberg; 2004:266–81.
-
- Bennell AP, Hu CM. Pollen morphology and taxonomy of lysimachia. Notes Royal Botanic Garden Edinburgh. 1983;40:425–458.
-
- Chen FH, Hu CM. Taxonomic and phytogeographic studies on Chinese species of lysimachia. Acta Phytotaxon Sin. 1979;17:21–56.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

