Neotropical bee microbiomes point to a fragmented social core and strong species-level effects
- PMID: 37452376
- PMCID: PMC10347802
- DOI: 10.1186/s40168-023-01593-z
Neotropical bee microbiomes point to a fragmented social core and strong species-level effects
Abstract
Background: Individuals that band together create new ecological opportunities for microorganisms. In vertical transmission, theory predicts a conserved microbiota within lineages, especially social bees. Bees exhibit solitary to social behavior among and/or within species, while life cycles can be annual or perennial. Bee nests may be used over generations or only once, and foraging ecology varies widely. To assess which traits are associated with bee microbiomes, we analyzed microbial diversity within solitary and social bees of Apidae, Colletidae, and Halictidae, three bee families in Panama's tropical forests. Our analysis considered the microbiome of adult gut contents replicated through time, localities, and seasons (wet and dry) and included bee morphology and comparison to abdominal (dissected) microbiota. Diversity and distribution of tropical bee microbes (TBM) within the corbiculate bee clade were emphasized.
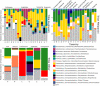
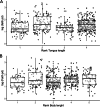
Results: We found the eusocial corbiculate bees tended to possess a more conserved gut microbiome, attributable to vertical transmission, but microbial composition varied among closely related species. Euglossine bees (or orchid bees), corbiculates with mainly solitary behavior, had more variable gut microbiomes. Their shorter-tongued and highly seasonal species displayed greater diversity, attributable to flower-visiting habits. Surprisingly, many stingless bees, the oldest corbiculate clade, lacked bacterial genera thought to predate eusociality, while several facultatively social, and solitary bee species possessed those bacterial taxa. Indeed, nearly all bee species displayed a range of affinities for single or multiple variants of the "socially associated" bacterial taxa, which unexpectedly demonstrated high sequence variation.
Conclusions: Taken together, these results call into question whether specific bacterial associates facilitate eusocial behavior, or are subsequently adopted, or indicate frequent horizontal transmission between perennial eusocial colonies and other social, facultatively social, and solitary bees. Video Abstract.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Wcislo WT. The roles of seasonality, host synchrony, and behaviour in the evolutions and distributions of nest parasites in Hymenoptera (Insecta), with special reference to bees (Apoidea) Biol Rev. 1987;62:515–543. doi: 10.1111/j.1469-185X.1987.tb01640.x. - DOI
-
- Roubik DW. Ecology and social organisation of bees. Encyclopedia of Life Sciences. 2012. 10.1002/9780470015902.a0023596.
-
- Hamilton WD. Kinship, recognition, disease, and intelligence: constraints of social evolution. 1987.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

