Recurrent allopolyploidizations diversify ecophysiological traits in marsh orchids (Dactylorhiza majalis s.l.)
- PMID: 37452724
- PMCID: PMC10947288
- DOI: 10.1111/mec.17070
Recurrent allopolyploidizations diversify ecophysiological traits in marsh orchids (Dactylorhiza majalis s.l.)
Abstract
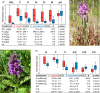
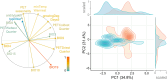
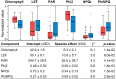
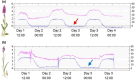
Whole-genome duplication has shaped the evolution of angiosperms and other organisms, and is important for many crops. Structural reorganization of chromosomes and repatterning of gene expression are frequently observed in allopolyploids, with physiological and ecological consequences. Recurrent origins from different parental populations are widespread among polyploids, resulting in an array of lineages that provide excellent models to uncover mechanisms of adaptation to divergent environments in early phases of polyploid evolution. We integrate here transcriptomic and ecophysiological comparative studies to show that sibling allopolyploid marsh orchid species (Dactylorhiza, Orchidaceae) occur in different habitats (low nutrient fens vs. meadows with mesic soils) and are characterized by a complex suite of intertwined, pronounced ecophysiological differences between them. We uncover distinct features in leaf elemental chemistry, light-harvesting, photoprotection, nutrient transport and stomata activity of the two sibling allopolyploids, which appear to match their specific ecologies, in particular soil chemistry differences at their native sites. We argue that the phenotypic divergence between the sibling allopolyploids has a clear genetic basis, generating ecological barriers that maintain distinct, independent lineages, despite pervasive interspecific gene flow. This suggests that recurrent origins of polyploids bring about a long-term potential to trigger and maintain functional and ecological diversity in marsh orchids and other groups.
Keywords: Dactylorhiza; allopolyploidy; differential expression; ecological differentiation; photosynthesis; soil chemistry.
© 2023 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Aagaard, S. M. D. , Sastad, S. M. , Greilhuber, J. , & Moen, A. A. (2005). Secondary hybrid zone between diploid Dactylorhiza incarnata subsp. cruenta and allotetraploid D. lapponica (Orchidaceae). Heredity, 94, 488–496. - PubMed
-
- Abbott, R. , Albach, D. , Ansell, S. , Arntzen, J. W. , Baird, S. J. E. , Bierne, N. , Boughman, J. , Brelsford, A. , Buerkle, C. A. , Buggs, R. , Butlin, R. K. , Dieckmann, U. , Eroukhmanoff, F. , Grill, A. , Cahan, S. H. , Hermansen, J. S. , Hewitt, G. , Hudson, A. G. , Jiggins, C. , … Zinner, D. (2013). Hybridization and speciation. Journal of Evolutionary Biology, 26(2), 229–246. - PubMed
-
- Ackerman, J. D. , Trejo‐Torres, J. C. , & Crespo‐Chuy, Y. (2007). Orchids of the West Indies: Predictability of diversity and endemism. Journal of Biogeography, 34, 779–786.
-
- Adams, K. L. , & Wendel, J. F. (2005). Novel patterns of gene expression in polyploid plants. Trends in Genetics, 21(10), 539–543. - PubMed
-
- Alexa, A. , & Rahnenfuhrer, J. (2018). topGO: Enrichment analysis for gene ontology. R Package Version 2.36.0.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

