Discovery of a glutathione utilization pathway in Francisella that shows functional divergence between environmental and pathogenic species
- PMID: 37453420
- PMCID: PMC10763578
- DOI: 10.1016/j.chom.2023.06.010
Discovery of a glutathione utilization pathway in Francisella that shows functional divergence between environmental and pathogenic species
Abstract
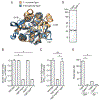
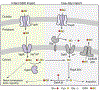
Glutathione (GSH) is an abundant metabolite within eukaryotic cells that can act as a signal, a nutrient source, or serve in a redox capacity for intracellular bacterial pathogens. For Francisella, GSH is thought to be a critical in vivo source of cysteine; however, the cellular pathways permitting GSH utilization by Francisella differ between strains and have remained poorly understood. Using genetic screening, we discovered a unique pathway for GSH utilization in Francisella. Whereas prior work suggested GSH catabolism initiates in the periplasm, the pathway we define consists of a major facilitator superfamily (MFS) member that transports intact GSH and a previously unrecognized bacterial cytoplasmic enzyme that catalyzes the first step of GSH degradation. Interestingly, we find that the transporter gene for this pathway is pseudogenized in pathogenic Francisella, explaining phenotypic discrepancies in GSH utilization among Francisella spp. and revealing a critical role for GSH in the environmental niche of these bacteria.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



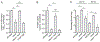
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

