LRRK2 phosphorylation status and kinase activity regulate (macro)autophagy in a Rab8a/Rab10-dependent manner
- PMID: 37454104
- PMCID: PMC10349885
- DOI: 10.1038/s41419-023-05964-0
LRRK2 phosphorylation status and kinase activity regulate (macro)autophagy in a Rab8a/Rab10-dependent manner
Abstract
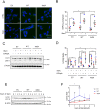
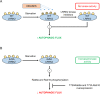
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most common genetic cause of Parkinson's disease (PD), with growing importance also for Crohn's disease and cancer. LRRK2 is a large and complex protein possessing both GTPase and kinase activity. Moreover, LRRK2 activity and function can be influenced by its phosphorylation status. In this regard, many LRRK2 PD-associated mutants display decreased phosphorylation of the constitutive phosphorylation cluster S910/S935/S955/S973, but the role of these changes in phosphorylation status with respect to LRRK2 physiological functions remains unknown. Here, we propose that the S910/S935/S955/S973 phosphorylation sites act as key regulators of LRRK2-mediated autophagy under both basal and starvation conditions. We show that quadruple LRRK2 phosphomutant cells (4xSA; S910A/S935A/S955A/S973A) have impaired lysosomal functionality and fail to induce and proceed with autophagy during starvation. In contrast, treatment with the specific LRRK2 kinase inhibitors MLi-2 (100 nM) or PF-06447475 (150 nM), which also led to decreased LRRK2 phosphorylation of S910/S935/S955/S973, did not affect autophagy. In explanation, we demonstrate that the autophagy impairment due to the 4xSA LRRK2 phospho-dead mutant is driven by its enhanced LRRK2 kinase activity. We show mechanistically that this involves increased phosphorylation of LRRK2 downstream targets Rab8a and Rab10, as the autophagy impairment in 4xSA LRRK2 cells is counteracted by expression of phosphorylation-deficient mutants T72A Rab8a and T73A Rab10. Similarly, reduced autophagy and decreased LRRK2 phosphorylation at the constitutive sites were observed in cells expressing the pathological R1441C LRRK2 PD mutant, which also displays increased kinase activity. These data underscore the relation between LRRK2 phosphorylation at its constitutive sites and the importance of increased LRRK2 kinase activity in autophagy regulation and PD pathology.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

