Investigation of multidrug-resistant plasmids from carbapenemase-producing Klebsiella pneumoniae clinical isolates from Pakistan
- PMID: 37455731
- PMCID: PMC10340517
- DOI: 10.3389/fmicb.2023.1192097
Investigation of multidrug-resistant plasmids from carbapenemase-producing Klebsiella pneumoniae clinical isolates from Pakistan
Abstract
Objectives: The study aim was to investigate multidrug-resistant (MDR) plasmids from a collection of 10 carbapenemase-producing Klebsiella pneumoniae clinical isolates identified within the same healthcare institution in Pakistan. Full characterization of the MDR plasmids including structure, typing characteristics, and AMR content as well as determination of their plasmid-based antimicrobial susceptibility profiles were carried out.
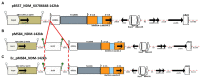
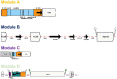
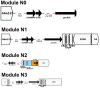
Methods: Plasmids were isolated from 10 clinical isolates of Klebsiella pneumoniae, and from a corresponding set of Escherichia coli transconjugants, then sequenced using Nanopore/Illumina technology to generate plasmid hybrid assemblies. Full characterization of MDR plasmids, including determination of next generation sequencing (NGS)-based AMR profiles, plasmid incompatibility groups, and types, was carried out. The structure of MDR plasmids was analyzed using the Galileo AMR platform. For E. coli transconjugants, the NGS-based AMR profiles were compared to NGS-predicted AMR phenotypes and conventional broth microdilution (BMD) antimicrobial susceptibility testing (AST) results.
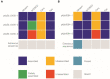
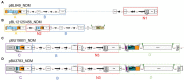
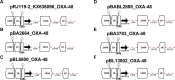
Results: All carbapenemase-producing K. pneumoniae isolates (carrying either blaNDM-1, or/and blaOXA-48) carried multiple AMR plasmids encoding 34 antimicrobial resistance genes (ARGs) conferring resistance to antimicrobials from 6 different classes. The plasmid incompatibility groups and types identified were: IncC (types 1 and 3), IncFIA (type 26) IncFIB, IncFII (types K1, K2, K7, and K9), IncHI1B, and IncL. None of the blaNDM-1 and blaESBL-plasmids identified in this study were previously described. Most blaNDM-1-plasmids shared identical AMR regions suggesting potential genetic material/plasmid exchange between K. pneumoniae isolates of this collection. The majority of NGS-based AMR profiles from the E. coli transconjugants correlated well with both NGS-based predicted and conventional AST results.
Conclusion: This study highlights the complexity and diversity of the plasmid-based genetic background of carbapenemase-producing clinical isolates from Pakistan. This study emphasizes the need for characterization of MDR plasmids to determine their complete molecular background and monitor AMR through plasmid transmission between multi-resistant bacterial pathogens.
Keywords: AMR data analysis; AMR determinants; Klebsiella pneumoniae; MDR plasmids; nanopore hybrid assemblies.
Copyright © 2023 Lascols, Cherney, Conley, Rishishwar, Crawford, Morse, Fisher, Anderson, Hodge, Pillai, Hughes, Khan and Sue.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

