Multi-year molecular quantification and 'omics analysis of Planktothrix-specific cyanophage sequences from Sandusky Bay, Lake Erie
- PMID: 37455749
- PMCID: PMC10343443
- DOI: 10.3389/fmicb.2023.1199641
Multi-year molecular quantification and 'omics analysis of Planktothrix-specific cyanophage sequences from Sandusky Bay, Lake Erie
Abstract
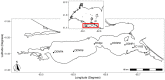
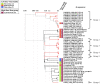
Introduction: Planktothrix agardhii is a microcystin-producing cyanobacterium found in Sandusky Bay, a shallow and turbid embayment of Lake Erie. Previous work in other systems has indicated that cyanophages are an important natural control factor of harmful algal blooms. Currently, there are few cyanophages that are known to infect P. agardhii, with the best-known being PaV-LD, a tail-less cyanophage isolated from Lake Donghu, China. Presented here is a molecular characterization of Planktothrix specific cyanophages in Sandusky Bay.
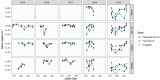
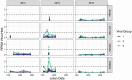
Methods and results: Putative Planktothrix-specific viral sequences from metagenomic data from the bay in 2013, 2018, and 2019 were identified by two approaches: homology to known phage PaV-LD, or through matching CRISPR spacer sequences with Planktothrix host genomes. Several contigs were identified as having viral signatures, either related to PaV-LD or potentially novel sequences. Transcriptomic data from 2015, 2018, and 2019 were also employed for the further identification of cyanophages, as well as gene expression of select viral sequences. Finally, viral quantification was tested using qPCR in 2015-2019 for PaV-LD like cyanophages to identify the relationship between presence and gene expression of these cyanophages. Notably, while PaV-LD like cyanophages were in high abundance over the course of multiple years (qPCR), transcriptomic analysis revealed only low levels of viral gene expression.
Discussion: This work aims to provide a broader understanding of Planktothrix cyanophage diversity with the goals of teasing apart the role of cyanophages in the control and regulation of harmful algal blooms and designing monitoring methodology for potential toxin-releasing lysis events.
Keywords: PaV-LD; Planktothrix agardhii; cyanophage; metagenome; metatranscriptome; qPCR.
Copyright © 2023 McKindles, Manes, Neudeck, McKay and Bullerjahn.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bullerjahn G. S., McKay R. M.. (2020a). Data from Sandusky Bay, Lake Erie from surveys conducted via Ohio Dept of natural resources watercraft from June to September 2018. Biological and chemical oceanography data management office (BCO-DMO). (version 1) version date 2019-02-07.
-
- Bullerjahn G. S., McKay R. M.. (2020b). Data from Sandusky Bay, Lake Erie from surveys conducted via Ohio Dept of natural resources watercraft from June to September 2019. Biological and chemical oceanography data management office (BCO-DMO). (version 1) version date 2020-06-09.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

