The genome sequence of the Loggerhead sea turtle, Caretta caretta Linnaeus 1758
- PMID: 37455852
- PMCID: PMC10338980
- DOI: 10.12688/f1000research.131283.2
The genome sequence of the Loggerhead sea turtle, Caretta caretta Linnaeus 1758
Abstract
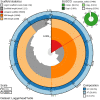
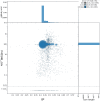
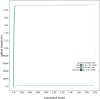
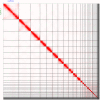
We present a genome assembly of Caretta caretta (the Loggerhead sea turtle; Chordata, Testudines, Cheloniidae), generated from genomic data from two unrelated females. The genome sequence is 2.13 gigabases in size. The assembly has a busco completion score of 96.1% and N50 of 130.95 Mb. The majority of the assembly is scaffolded into 28 chromosomal representations with a remaining 2% of the assembly being excluded from these.
Keywords: Caretta caretta; Loggerhead sea turtle; chromosomal; genome sequence; reptile.
Copyright: © 2023 Chang G et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
References
-
- Casale P, Tucker A: Caretta caretta (amended version of 2015 assessment). IUCN red list of threatened species. 2015. 10.2305/iucn.uk.2017-2.rlts.t3897a119333622.en - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

