In-silico, evolutionary, and functional analysis of CHUP1 and its related proteins in Bienertia sinuspersici-a comparative study across C3, C4, CAM, and SCC4 model plants
- PMID: 37456874
- PMCID: PMC10348308
- DOI: 10.7717/peerj.15696
In-silico, evolutionary, and functional analysis of CHUP1 and its related proteins in Bienertia sinuspersici-a comparative study across C3, C4, CAM, and SCC4 model plants
Abstract
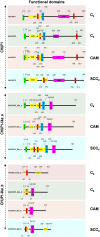
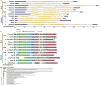
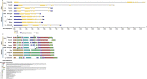
Single-cell C4 (SCC4) plants with bienertioid anatomy carry out photosynthesis in a single cell. Chloroplast movement is the underlying phenomenon, where chloroplast unusual positioning 1 (CHUP1) plays a key role. This study aimed to characterize CHUP1 and CHUP1-like proteins in an SCC4 photosynthetic plant, Bienertia sinuspersici. Also, a comparative analysis of SCC4 CHUP1 was made with C3, C4, and CAM model plants including an extant basal angiosperm, Amborella. The CHUP1 gene exists as a single copy from the basal angiosperms to SCC4 plants. Our analysis identified that Chenopodium quinoa, a recently duplicated allotetraploid, has two copies of CHUP1. In addition, the numbers of CHUP1-like and its associated proteins such as CHUP1-like_a, CHUP1-like_b, HPR, TPR, and ABP varied between the species. Hidden Markov Model analysis showed that the gene size of CHUP1-like_a and CHUP1-like_b of SCC4 species, Bienertia, and Suaeda were enlarged than other plants. Also, we identified that CHUP1-like_a and CHUP1-like_b are absent in Arabidopsis and Amborella, respectively. Motif analysis identified several conserved and variable motifs based on the orders (monocot and dicot) as well as photosynthetic pathways. For instance, CAM plants such as pineapple and cactus shared certain motifs of CHUP1-like_a irrespective of their distant phylogenetic relationship. The free ratio model showed that CHUP1 maintained purifying selection, whereas CHUP1-like_a and CHUP1-like_b have adaptive functions between SCC4 plants and quinoa. Similarly, rice and maize branches displayed functional diversification on CHUP1-like_b. Relative gene expression data showed that during the subcellular compartmentalization process of Bienertia, CHUP1 and actin-binding proteins (ABP) genes showed a similar pattern of expression. Altogether, the results of this study provide insight into the evolutionary and functional details of CHUP1 and its associated proteins in the development of the SCC4 system in comparison with other C3, C4, and CAM model plants.
Keywords: Bienertia sinuspersici; CHUP1 protein; CHUP1-like proteins; Phylogenetic analysis; Single-Cell C4 plants; Subcellular compartmentalization.
©2023 Won et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
Similar articles
-
Comparative transcriptome analysis of emerging young and mature leaves of Bienertia sinuspersici, a single-cell C4 plant.PeerJ. 2025 Apr 29;13:e19282. doi: 10.7717/peerj.19282. eCollection 2025. PeerJ. 2025. PMID: 40321813 Free PMC article.
-
Comparative transcriptomics reveals the role of altered energy metabolism in the establishment of single-cell C4 photosynthesis in Bienertia sinuspersici.Front Plant Sci. 2023 Jul 5;14:1202521. doi: 10.3389/fpls.2023.1202521. eCollection 2023. Front Plant Sci. 2023. PMID: 37476170 Free PMC article.
-
Evolution of RLSB, a nuclear-encoded S1 domain RNA binding protein associated with post-transcriptional regulation of plastid-encoded rbcL mRNA in vascular plants.BMC Evol Biol. 2016 Jun 29;16(1):141. doi: 10.1186/s12862-016-0713-1. BMC Evol Biol. 2016. PMID: 27356975 Free PMC article.
-
One decade after the discovery of single-cell C4 species in terrestrial plants: what did we learn about the minimal requirements of C4 photosynthesis?Photosynth Res. 2014 Feb;119(1-2):169-80. doi: 10.1007/s11120-013-9810-9. Epub 2013 Mar 14. Photosynth Res. 2014. PMID: 23494362 Review.
-
Temperature response of photosynthesis in C3, C4, and CAM plants: temperature acclimation and temperature adaptation.Photosynth Res. 2014 Feb;119(1-2):101-17. doi: 10.1007/s11120-013-9874-6. Epub 2013 Jun 26. Photosynth Res. 2014. PMID: 23801171 Review.
Cited by
-
Genome-wide identification of a novel Na+ transporter from Bienertia sinuspersici and overexpression of BsHKT1;2 improved salt tolerance in Brassica rapa.Front Plant Sci. 2023 Dec 12;14:1302315. doi: 10.3389/fpls.2023.1302315. eCollection 2023. Front Plant Sci. 2023. PMID: 38192689 Free PMC article.
References
-
- Akhani H, Barroca J, Koteeva N, Voznesenskaya E, Franceschi V, Edwards G, Ghaffari SM, Ziegler H. Bienertia sinuspersici (Chenopodiaceae): a new species from Southwest Asia and discovery of a third terrestrial C4 plant without Kranz anatomy. Systematic Botany. 2005;30:290–301. doi: 10.1600/0363644054223684. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

