Direct detection of drug-resistant Mycobacterium tuberculosis using targeted next generation sequencing
- PMID: 37457262
- PMCID: PMC10340549
- DOI: 10.3389/fpubh.2023.1206056
Direct detection of drug-resistant Mycobacterium tuberculosis using targeted next generation sequencing
Abstract
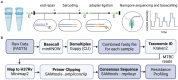
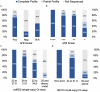
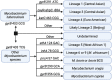
Mycobacterium tuberculosis complex (MTBC) infections are treated with combinations of antibiotics; however, these regimens are not as efficacious against multidrug and extensively drug resistant MTBC. Phenotypic (growth-based) drug susceptibility testing on slow growing bacteria like MTBC requires many weeks to months to complete, whereas sequencing-based approaches can predict drug resistance (DR) with reduced turnaround time. We sought to develop a multiplexed, targeted next generation sequencing (tNGS) assay that can predict DR and can be performed directly on clinical respiratory specimens. A multiplex PCR was designed to amplify a group of thirteen full-length genes and promoter regions with mutations known to be involved in resistance to first- and second-line MTBC drugs. Long-read amplicon libraries were sequenced with Oxford Nanopore Technologies platforms and high-confidence resistance mutations were identified in real-time using an in-house developed bioinformatics pipeline. Sensitivity, specificity, reproducibility, and accuracy of the tNGS assay was assessed as part of a clinical validation study. In total, tNGS was performed on 72 primary specimens and 55 MTBC-positive cultures and results were compared to clinical whole genome sequencing (WGS) performed on paired patient cultures. Complete or partial susceptibility profiles were generated from 82% of smear positive primary specimens and the resistance mutations identified by tNGS were 100% concordant with WGS. In addition to performing tNGS on primary clinical samples, this assay can be used to sequence MTBC cultures mixed with other mycobacterial species that would not yield WGS results. The assay can be effectively implemented in a clinical/diagnostic laboratory with a two to three day turnaround time and, even if batched weekly, tNGS results are available on average 15 days earlier than culture-derived WGS results. This study demonstrates that tNGS can reliably predict MTBC drug resistance directly from clinical specimens or cultures and provide critical information in a timely manner for the appropriate treatment of patients with DR tuberculosis.
Keywords: drug susceptibility; mycobacterium; nanopore; resistance; targeted sequencing; tuberculosis.
Copyright © 2023 Murphy, Smith, Lapierre, Shea, Patel, Halse, Dickinson, Escuyer, Rowlinson and Musser.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- World Health Organization . Annual report of tuberculosis. Annual Global TB Report of WHO (2022) 8:1–68. Available at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... (Accessed March 13, 2023).
-
- Doulla BE, Squire SB, MacPherson E, Ngadaya ES, Mutayoba BK, Langley I. Routine surveillance for the identification of drug resistant tuberculosis in Tanzania: a cross-sectional study of stakeholders’ perceptions. PLoS One. (2019) 14:e0212421. doi: 10.1371/JOURNAL.PONE.0212421, PMID: - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

