Revisiting ESKAPE Pathogens: virulence, resistance, and combating strategies focusing on quorum sensing
- PMID: 37457962
- PMCID: PMC10339816
- DOI: 10.3389/fcimb.2023.1159798
Revisiting ESKAPE Pathogens: virulence, resistance, and combating strategies focusing on quorum sensing
Abstract
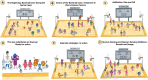
The human-bacterial association is long-known and well-established in terms of both augmentations of human health and attenuation. However, the growing incidents of nosocomial infections caused by the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp.) call for a much deeper understanding of these organisms. Adopting a holistic approach that includes the science of infection and the recent advancements in preventing and treating infections is imperative in designing novel intervention strategies against ESKAPE pathogens. In this regard, this review captures the ingenious strategies commissioned by these master players, which are teamed up against the defenses of the human team, that are equally, if not more, versatile and potent through an analogy. We have taken a basketball match as our analogy, dividing the human and bacterial species into two teams playing with the ball of health. Through this analogy, we make the concept of infectious biology more accessible.
Keywords: ESKAPE; antimicrobial resistance; biofilm; quorum sensing; virulence.
Copyright © 2023 Venkateswaran, Vasudevan, David, Shaktivel, Shanmugam, Neelakantan and Solomon.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

