Ionizable Lipid Nanoparticles for Therapeutic Base Editing of Congenital Brain Disease
- PMID: 37458484
- PMCID: PMC11025390
- DOI: 10.1021/acsnano.3c02268
Ionizable Lipid Nanoparticles for Therapeutic Base Editing of Congenital Brain Disease
Abstract
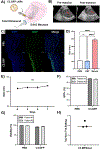
Delivery of mRNA-based therapeutics to the perinatal brain holds great potential in treating congenital brain diseases. However, nonviral delivery platforms that facilitate nucleic acid delivery in this environment have yet to be rigorously studied. Here, we screen a diverse library of ionizable lipid nanoparticles (LNPs) via intracerebroventricular (ICV) injection in both fetal and neonatal mice and identify an LNP formulation with greater functional mRNA delivery in the perinatal brain than an FDA-approved industry standard LNP. Following in vitro optimization of the top-performing LNP (C3 LNP) for codelivery of an adenine base editing platform, we improve the biochemical phenotype of a lysosomal storage disease in the neonatal mouse brain, exhibit proof-of-principle mRNA brain transfection in vivo in a fetal nonhuman primate model, and demonstrate the translational potential of C3 LNPs ex vivo in human patient-derived brain tissues. These LNPs may provide a clinically translatable platform for in utero and postnatal mRNA therapies including gene editing in the brain.
Keywords: congenital brain disease; fetal gene therapy; gene editing; ionizable lipid nanoparticles; mRNA delivery.
Conflict of interest statement
Figures
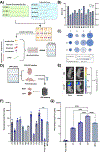
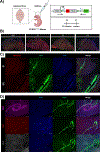
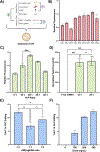
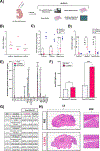
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

