Complexity of Guanine Quadruplex Unfolding Pathways Revealed by Atomistic Pulling Simulations
- PMID: 37458574
- PMCID: PMC10428220
- DOI: 10.1021/acs.jcim.3c00171
Complexity of Guanine Quadruplex Unfolding Pathways Revealed by Atomistic Pulling Simulations
Abstract
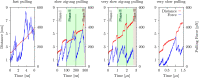
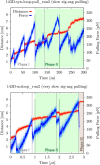
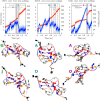
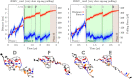
Guanine quadruplexes (GQs) are non-canonical nucleic acid structures involved in many biological processes. GQs formed in single-stranded regions often need to be unwound by cellular machinery, so their mechanochemical properties are important. Here, we performed steered molecular dynamics simulations of human telomeric GQs to study their unfolding. We examined four pulling regimes, including a very slow setup with pulling velocity and force load accessible to high-speed atomic force microscopy. We identified multiple factors affecting the unfolding mechanism, i.e.,: (i) the more the direction of force was perpendicular to the GQ channel axis (determined by GQ topology), the more the base unzipping mechanism happened, (ii) the more parallel the direction of force was, GQ opening and cross-like GQs were more likely to occur, (iii) strand slippage mechanism was possible for GQs with an all-anti pattern in a strand, and (iv) slower pulling velocity led to richer structural dynamics with sampling of more intermediates and partial refolding events. We also identified that a GQ may eventually unfold after a force drop under forces smaller than those that the GQ withstood before the drop. Finally, we found out that different unfolding intermediates could have very similar chain end-to-end distances, which reveals some limitations of structural interpretations of single-molecule spectroscopic data.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Mechanical Stability and Unfolding Pathways of Parallel Tetrameric G-Quadruplexes Probed by Pulling Simulations.J Chem Inf Model. 2024 May 13;64(9):3896-3911. doi: 10.1021/acs.jcim.4c00227. Epub 2024 Apr 17. J Chem Inf Model. 2024. PMID: 38630447 Free PMC article.
-
Folding of guanine quadruplex molecules-funnel-like mechanism or kinetic partitioning? An overview from MD simulation studies.Biochim Biophys Acta Gen Subj. 2017 May;1861(5 Pt B):1246-1263. doi: 10.1016/j.bbagen.2016.12.008. Epub 2016 Dec 13. Biochim Biophys Acta Gen Subj. 2017. PMID: 27979677 Review.
-
Force-induced unfolding of human telomeric G-quadruplex: a steered molecular dynamics simulation study.Biochem Biophys Res Commun. 2009 Jan 30;379(1):70-5. doi: 10.1016/j.bbrc.2008.12.006. Epub 2008 Dec 13. Biochem Biophys Res Commun. 2009. PMID: 19073141
-
Same fold, different properties: polarizable molecular dynamics simulations of telomeric and TERRA G-quadruplexes.Nucleic Acids Res. 2020 Jan 24;48(2):561-575. doi: 10.1093/nar/gkz1154. Nucleic Acids Res. 2020. PMID: 31807754 Free PMC article.
-
G-quadruplex dynamics.Biochim Biophys Acta Proteins Proteom. 2017 Nov;1865(11 Pt B):1544-1554. doi: 10.1016/j.bbapap.2017.06.012. Epub 2017 Jun 20. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 28642152 Review.
Cited by
-
Mechanical Stability and Unfolding Pathways of Parallel Tetrameric G-Quadruplexes Probed by Pulling Simulations.J Chem Inf Model. 2024 May 13;64(9):3896-3911. doi: 10.1021/acs.jcim.4c00227. Epub 2024 Apr 17. J Chem Inf Model. 2024. PMID: 38630447 Free PMC article.
-
Computer Folding of Parallel DNA G-Quadruplex: Hitchhiker's Guide to the Conformational Space.J Comput Chem. 2025 Jan 5;46(1):e27535. doi: 10.1002/jcc.27535. J Comput Chem. 2025. PMID: 39653644 Free PMC article.
References
-
- Lee W. T. C.; Yin Y.; Morten M. J.; Tonzi P.; Gwo P. P.; Odermatt D. C.; Modesti M.; Cantor S. B.; Gari K.; Huang T. T.; et al. Single-Molecule Imaging Reveals Replication Fork Coupled Formation of G-Quadruplex Structures Hinders Local Replication Stress Signaling. Nat. Commun. 2021, 12, 2525.10.1038/s41467-021-22830-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

