Therapeutic Switching of Rafoxanide: a New Approach To Fighting Drug-Resistant Bacteria and Fungi
- PMID: 37458598
- PMCID: PMC10433953
- DOI: 10.1128/spectrum.02679-22
Therapeutic Switching of Rafoxanide: a New Approach To Fighting Drug-Resistant Bacteria and Fungi
Abstract
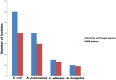
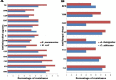
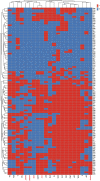
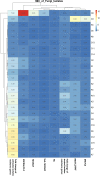
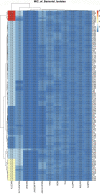
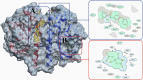
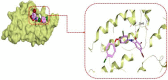
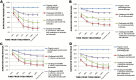
Control and management of life-threatening bacterial and fungal infections are a global health challenge. Despite advances in antimicrobial therapies, treatment failures for resistant bacterial and fungal infections continue to increase. We aimed to repurpose the anthelmintic drug rafoxanide for use with existing therapeutic drugs to increase the possibility of better managing infection and decrease treatment failures. For this purpose, we evaluated the antibacterial and antifungal potential of rafoxanide. Notably, 70% (70/100) of bacterial isolates showed multidrug resistance (MDR) patterns, with higher prevalence among human isolates (73.5% [50/68]) than animal ones (62.5% [20/32]). Moreover, 22 fungal isolates (88%) were MDR and were more prevalent among animal (88.9%) than human (87.5%) sources. We observed alarming MDR patterns among bacterial isolates, i.e., Klebsiella pneumoniae (75% [30/40; 8 animal and 22 human]) and Escherichia coli (66% [40/60; 12 animal and 28 human]), and fungal isolates, i.e., Candida albicans (86.7% [13/15; 4 animal and 9 human]) and Aspergillus fumigatus (90% [9/10; 4 animal and 5 human]), that were resistant to at least one agent in three or more different antimicrobial classes. Rafoxanide had antibacterial and antifungal activities, with minimal inhibitory concentration (MICs) ranging from 2 to 128 μg/mL. Rafoxanide at sub-MICs downregulated the mRNA expression of resistance genes, including E. coli and K. pneumoniae blaCTX-M-1, blaTEM-1, blaSHV, MOX, and DHA, C. albicans ERG11, and A. fumigatus cyp51A. We noted the improvement in the activity of β-lactam and antifungal drugs upon combination with rafoxanide. This was apparent in the reduction in the MICs of cefotaxime and fluconazole when these drugs were combined with sub-MIC levels of rafoxanide. There was obvious synergism between rafoxanide and cefotaxime against all E. coli and K. pneumoniae isolates (fractional inhibitory concentration index [FICI] values ≤ 0.5). Accordingly, there was a shift in the patterns of resistance of 16.7% of E. coli and 22.5% of K. pneumoniae isolates to cefotaxime and those of 63.2% of C. albicans and A. fumigatus isolates to fluconazole when the isolates were treated with sub-MICs of rafoxanide. These results were confirmed by in silico and mouse protection assays. Based on the in silico study, one possible explanation for how rafoxanide reduced bacterial resistance is through its inhibitory effects on bacterial and fungal histidine kinase enzymes. In short, rafoxanide exhibited promising results in overcoming bacterial and fungal drug resistance. IMPORTANCE The drug repurposing strategy is an alternative approach to reducing drug development timelines with low cost, especially during outbreaks of disease caused by drug-resistant pathogens. Rafoxanide can disrupt the abilities of bacterial and fungal cells to adapt to stress conditions. The coadministration of antibiotics with rafoxanide can prevent the failure of treatment of both resistant bacteria and fungi, as the resistant pathogens could be made sensitive upon treatment with rafoxanide. From our findings, we anticipate that pharmaceutical companies will be able to utilize new combinations against resistant pathogens.
Keywords: in silico; mouse protection; rafoxanide; repurposing; resistance; resistance gene downregulation; treatment failure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi:10.1111/j.1469-0691.2011.03570.x. - DOI - PubMed
-
- Ammar AM, Abd El-Hamid MI, El-Malt RMS, Azab DS, Albogami S, Al-Sanea MM, Soliman WE, Ghoneim MM, Bendary MM. 2021. Molecular detection of fluoroquinolone resistance among multidrug-, extensively drug- and pan-drug-resistant Campylobacter species in Egypt. Antibiotics 10:1342. doi:10.3390/antibiotics10111342. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

