The Energetic Landscape of Catch Bonds in TCR Interfaces
- PMID: 37459192
- PMCID: PMC10361606
- DOI: 10.4049/jimmunol.2300121
The Energetic Landscape of Catch Bonds in TCR Interfaces
Abstract
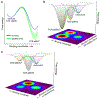
Recognition of peptide/MHC complexes by αβ TCRs has traditionally been viewed through the lens of conventional receptor-ligand theory. Recent work, however, has shown that TCR recognition and T cell signaling can be profoundly influenced and tuned by mechanical forces. One outcome of applied force is the catch bond, where TCR dissociation rates decrease (half-lives increase) when limited force is applied. Although catch bond behavior is believed to be widespread in biology, its counterintuitive nature coupled with the difficulties of describing mechanisms at the structural level have resulted in considerable mystique. In this review, we demonstrate that viewing catch bonds through the lens of energy landscapes, barriers, and the ensuing reaction rates can help demystify catch bonding and provide a foundation on which atomic-level TCR catch bond mechanisms can be built.
Copyright © 2023 by The American Association of Immunologists, Inc.
Figures
References
-
- Holler PD, and Kranz DM. 2003. Quantitative Analysis of the Contribution of TCR/pepMHC Affinity and CD8 to T Cell Activation. Immunity 18: 255–264. - PubMed
-
- Spear TT, Wang Y, Foley KC, Murray DC, Scurti GM, Simms PE, Garrett-Mayer E, Hellman LM, Baker BM, and Nishimura MI. 2017. Critical biological parameters modulate affinity as a determinant of function in T-cell receptor gene-modified T-cells. Cancer Immunology, Immunotherapy 66: 1411–1424. - PMC - PubMed
-
- Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, and Nathenson SG. 2001. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol 2: 229–234. - PubMed
-
- Baker BM, Gagnon SJ, Biddison WE, and Wiley DC. 2000. Conversion of a T Cell Antagonist into an Agonist by Repairing a Defect in the TCR/Peptide/MHC Interface. Implications for TCR Signaling. Immunity 13: 475–484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

