NRF2 activators inhibit influenza A virus replication by interfering with nucleo-cytoplasmic export of viral RNPs in an NRF2-independent manner
- PMID: 37459366
- PMCID: PMC10374058
- DOI: 10.1371/journal.ppat.1011506
NRF2 activators inhibit influenza A virus replication by interfering with nucleo-cytoplasmic export of viral RNPs in an NRF2-independent manner
Abstract
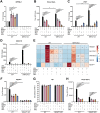
In addition to antioxidative and anti-inflammatory properties, activators of the cytoprotective nuclear factor erythroid-2-like-2 (NRF2) signaling pathway have antiviral effects, but the underlying antiviral mechanisms are incompletely understood. We evaluated the ability of the NRF2 activators 4-octyl itaconate (4OI), bardoxolone methyl (BARD), sulforaphane (SFN), and the inhibitor of exportin-1 (XPO1)-mediated nuclear export selinexor (SEL) to interfere with influenza virus A/Puerto Rico/8/1934 (H1N1) infection of human cells. All compounds reduced viral titers in supernatants from A549 cells and vascular endothelial cells in the order of efficacy SEL>4OI>BARD = SFN, which correlated with their ability to prevent nucleo-cytoplasmic export of viral nucleoprotein and the host cell protein p53. In contrast, intracellular levels of viral HA mRNA and nucleocapsid protein (NP) were unaffected. Knocking down mRNA encoding KEAP1 (the main inhibitor of NRF2) or inactivating the NFE2L2 gene (which encodes NRF2) revealed that physiologic NRF2 signaling restricts IAV replication. However, the antiviral effect of all compounds was NRF2-independent. Instead, XPO1 knock-down greatly reduced viral titers, and incubation of Calu3 cells with an alkynated 4OI probe demonstrated formation of a covalent complex with XPO1. Ligand-target modelling predicted covalent binding of all three NRF2 activators and SEL to the active site of XPO1 involving the critical Cys528. SEL and 4OI manifested the highest binding energies, whereby the 4-octyl tail of 4OI interacted extensively with the hydrophobic groove of XPO1, which binds nuclear export sequences on cargo proteins. Conversely, SEL as well as the three NRF2 activators were predicted to covalently bind the functionally critical Cys151 in KEAP1. Blocking XPO1-mediated nuclear export may, thus, constitute a "noncanonical" mechanism of anti-influenza activity of electrophilic NRF2 activators that can interact with similar cysteine environments at the active sites of XPO1 and KEAP1. Considering the importance of XPO1 function to a variety of pathogenic viruses, compounds that are optimized to inhibit both targets may constitute an important class of broadly active host-directed treatments that embody anti-inflammatory, cytoprotective, and antiviral properties.
Copyright: © 2023 Waqas et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Olagnier D, Farahani E, Thyrsted J, Blay-Cadanet J, Herengt A, Idorn M, et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat Commun. 2020;11(1):4938. Epub 2020/10/04. doi: 10.1038/s41467-020-18764-3 . - DOI - PMC - PubMed
-
- Chen WC, Huang CH, Liu W, Lee JC. Sulforaphane suppresses dengue virus replication by inhibition of dengue protease and enhancement of antiviral interferon response through Nrf2-mediated heme oxygenase-1 induction. Antiviral Res. 2022;207:105400. Epub 2022/09/03. doi: 10.1016/j.antiviral.2022.105400 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

