Analysis of ATG4C function in vivo
- PMID: 37459465
- PMCID: PMC10549197
- DOI: 10.1080/15548627.2023.2234799
Analysis of ATG4C function in vivo
Abstract
ATG4 (autophagy related 4 cysteine peptidase); ATG4A (autophagy related 4A cysteine peptidase); ATG4B (autophagy related 4B cysteine peptidase); ATG4C (autophagy related 4C cysteine peptidase); ATG4D (autophagy related 4D cysteine peptidase); Atg8 (autophagy related 8); GABARAP (GABA type A receptor-associated protein); GABARAPL1(GABA type A receptor-associated protein like 1); GABARAPL2 (GABA type A receptor-associated protein like 2); MAP1LC3A/LC3A (microtubule associated protein 1 light chain 3 alpha); MAP1LC3B/LC3B (microtubule associated protein 1 light chain 3 beta); mATG8 (mammalian Atg8); PE (phosphatidylethanolamine); PS (phosphatydylserine); SQSTM1/p62 (sequestosome 1).
Keywords: Animal models; GABARAP; LC3; autophagy; fibrosarcoma; lymphocyte.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

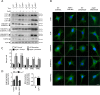


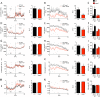
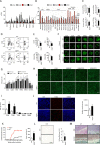
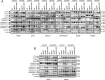

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
