OBOX regulates mouse zygotic genome activation and early development
- PMID: 37459895
- PMCID: PMC10528489
- DOI: 10.1038/s41586-023-06428-3
OBOX regulates mouse zygotic genome activation and early development
Abstract
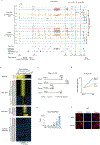
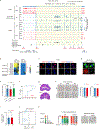
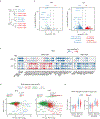
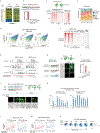
Zygotic genome activation (ZGA) activates the quiescent genome to enable the maternal-to-zygotic transition1,2. However, the identity of transcription factors that underlie mammalian ZGA in vivo remains elusive. Here we show that OBOX, a PRD-like homeobox domain transcription factor family (OBOX1-OBOX8)3-5, are key regulators of mouse ZGA. Mice deficient for maternally transcribed Obox1/2/5/7 and zygotically expressed Obox3/4 had a two-cell to four-cell arrest, accompanied by impaired ZGA. The Obox knockout defects could be rescued by restoring either maternal and zygotic OBOX, which suggests that maternal and zygotic OBOX redundantly support embryonic development. Chromatin-binding analysis showed that Obox knockout preferentially affected OBOX-binding targets. Mechanistically, OBOX facilitated the 'preconfiguration' of RNA polymerase II, as the polymerase relocated from the initial one-cell binding targets to ZGA gene promoters and distal enhancers. Impaired polymerase II preconfiguration in Obox mutants was accompanied by defective ZGA and chromatin accessibility transition, as well as aberrant activation of one-cell polymerase II targets. Finally, ectopic expression of OBOX activated ZGA genes and MERVL repeats in mouse embryonic stem cells. These data thus demonstrate that OBOX regulates mouse ZGA and early embryogenesis.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures











Comment in
-
Sleeping embryonic genomes are awoken by OBOX proteins.Nature. 2023 Aug;620(7976):956-957. doi: 10.1038/d41586-023-01618-5. Nature. 2023. PMID: 37460688 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

