Intermolecular electron transfer in radical SAM enzymes as a new paradigm for reductive activation
- PMID: 37460016
- PMCID: PMC10470005
- DOI: 10.1016/j.jbc.2023.105058
Intermolecular electron transfer in radical SAM enzymes as a new paradigm for reductive activation
Abstract
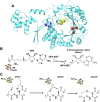
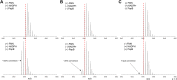
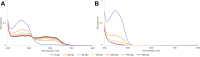
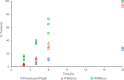
Radical S-adenosyl-L-methionine (rSAM) enzymes bind one or more Fe-S clusters and catalyze transformations that produce complex and structurally diverse natural products. One of the clusters, a 4Fe-4S cluster, binds and reductively cleaves SAM to generate the 5'-deoxyadenosyl radical, which initiates the catalytic cycle by H-atom transfer from the substrate. The role(s) of the additional auxiliary Fe-S clusters (ACs) remains largely enigmatic. The rSAM enzyme PapB catalyzes the formation of thioether cross-links between the β-carbon of an Asp and a Cys thiolate found in the PapA peptide. One of the two ACs in the protein binds to the substrate thiol where, upon formation of a thioether bond, one reducing equivalent is returned to the protein. However, for the next catalytic cycle to occur, the protein must undergo an electronic state isomerization, returning the electron to the SAM-binding cluster. Using a series of iron-sulfur cluster deletion mutants, our data support a model whereby the isomerization is an obligatorily intermolecular electron transfer event that can be mediated by redox active proteins or small molecules, likely via the second AC in PapB. Surprisingly, a mixture of FMN and NADPH is sufficient to support both the reductive and the isomerization steps. These findings lead to a new paradigm involving intermolecular electron transfer steps in the activation of rSAM enzymes that require multiple iron-sulfur clusters for turnover. The implications of these results for the biological activation of rSAM enzymes are discussed.
Keywords: S-adenosyl-L-methionine (SAM); electron transfer; enzymatic activation; enzyme mechanism; iron–sulfur protein; radical SAM.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest V. B. and K. A. S. E. have disclosed the results to the University of Utah, which holds patent interests in the findings.
Figures



References
-
- Oberg N., Precord T.W., Mitchell D.A., Gerlt J.A. RadicalSAM.org: a resource to interpret sequence-function space and discover new radical SAM enzyme chemistry. ACS Bio. Med. Chem. Au. 2022;2:22–35. - PMC - PubMed
-
- Sofia H.J., Chen G., Hetzler B.G., Reyes-Spindola J.F., Miller N.E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucl. Acids Res. 2001;29:1097. - PMC - PubMed
-
- Krebs C., Broderick W.E., Henshaw T.F., Broderick J.B., Huynh B.H. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating anzyme: a Mössbauer spectroscopic study. J. Am. Chem. Soc. 2002;124:912–913. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

