RelB-deficient autoinflammatory pathology presents as interferonopathy, but in mice is interferon-independent
- PMID: 37460023
- PMCID: PMC10858800
- DOI: 10.1016/j.jaci.2023.06.024
RelB-deficient autoinflammatory pathology presents as interferonopathy, but in mice is interferon-independent
Abstract
Background: Autoimmune diseases are leading causes of ill health and morbidity and have diverse etiology. Two signaling pathways are key drivers of autoimmune pathology, interferon and nuclear factor-κB (NF-κB)/RelA, defining the 2 broad labels of interferonopathies and relopathies. Prior work has established that genetic loss of function of the NF-κB subunit RelB leads to autoimmune and inflammatory pathology in mice and humans.
Objective: We sought to characterize RelB-deficient autoimmunity by unbiased profiling of the responses of immune sentinel cells to stimulus and to determine the functional role of dysregulated gene programs in the RelB-deficient pathology.
Methods: Transcriptomic profiling was performed on fibroblasts and dendritic cells derived from patients with RelB deficiency and knockout mice, and transcriptomic responses and pathology were assessed in mice deficient in both RelB and the type I interferon receptor.
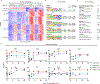
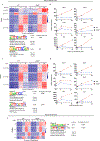
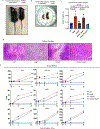
Results: We found that loss of RelB in patient-derived fibroblasts and mouse myeloid cells results in elevated induction of hundreds of interferon-stimulated genes. Removing hyperexpression of the interferon-stimulated gene program did not ameliorate the autoimmune pathology of RelB knockout mice. Instead, we found that RelB suppresses a different set of inflammatory response genes in a manner that is independent of interferon signaling but associated with NF-κB binding motifs.
Conclusion: Although transcriptomic profiling would describe RelB-deficient autoimmune disease as an interferonopathy, the genetic evidence indicates that the pathology in mice is interferon-independent.
Keywords: RelB; autoimmunity; dendritic cells; inflammation; interferonopathy; relopathy.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest:
The authors declare that research in this study was conducted in the absence of a conflict of interest.
Figures

References
-
- Committee for the Assessment of NIH Research on Autoimmune Diseases, Board on Population Health and Public Health Practice, Health and Medicine Division, National Academies of Sciences, Engineering, and Medicine. Enhancing NIH Research on Autoimmune Disease [Internet] Washington (DC): National Academies Press (US); 2022. [cited 2022 Nov 2]. (The National Academies Collection: Reports funded by National Institutes of Health). Available from: http://www.ncbi.nlm.nih.gov/books/NBK580299/ - PubMed
-
- Savic S, Coe J, Laws P. Autoinflammation: Interferonopathies and Other Autoinflammatory Diseases. J Invest Dermatol 2022. Mar;142(3 Pt B):781–92. - PubMed
-
- Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium - PubMed [Internet] [cited 2022 Nov 2]. Available from: https://pubmed.ncbi.nlm.nih.gov/9288758/ - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

